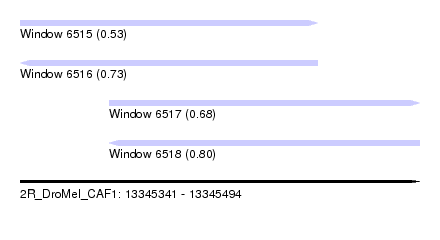
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,345,341 – 13,345,494 |
| Length | 153 |
| Max. P | 0.795793 |

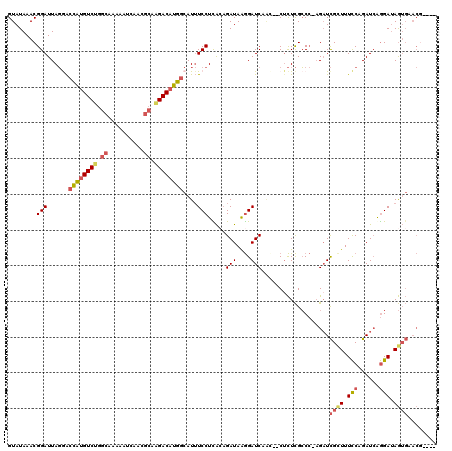
| Location | 13,345,341 – 13,345,455 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 83.31 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -20.20 |
| Energy contribution | -20.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
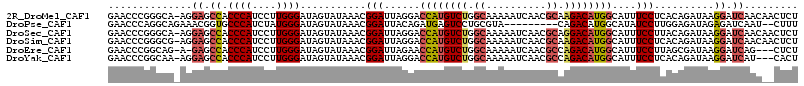
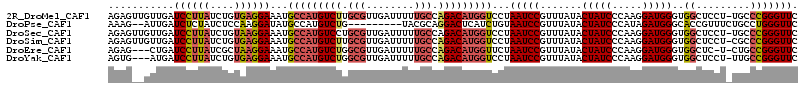
>2R_DroMel_CAF1 13345341 114 + 20766785 GAACCCGGGCA-AGGAGCCACCCAUCCUUGGGAUAGUAUAAACGGAUUAGGACCAUGUCUGGCAAAAAUCAACGCAAGACAUGGCAUUUCCUCACAGAUAAGGAUCAACAACUCU ....((((((.-....))).((((....))))..........))).....(.((((((((.((..........)).)))))))))...((((........))))........... ( -31.00) >DroPse_CAF1 42349 104 + 1 GAACCCAGGCAGAAACGGUGCCCAUCUAUGGGAUAGUAUAAACGGAUUACAGAUGAGUCCUGCGUA---------CAGACAUGGCAUAUCCUUGGAGAUAGAGAUCAAU--CUUU ....(((((..((....((((((((((.(((....((....))...))).))))).(((.((....---------)))))..))))).)))))))((((.(....).))--)).. ( -20.10) >DroSec_CAF1 44597 114 + 1 GAACCCGGGCA-AGGAGCCACCCAUCCUUGGGAUAGUAUAAACGGAUUAGGACCAUGUCUGGCAAAAAUCAACGCAGGACAUGGCAUUUCCUUACAGAUAAGGAUCAACAACUCU ....((((((.-....))).((((....))))..........))).....(.((((((((.((..........)).)))))))))...((((((....))))))........... ( -33.10) >DroSim_CAF1 46583 114 + 1 GAACCCGGGCG-AGGAGCCACCCAUCCUUGGGAUAGUAUAAACGGAUUAGGACCAUGUCUGGCAAAAAUCAACGCAAGACAUGGCAUUUCCUCACAGAUAAGGAUCAACAACUCU ....((((((.-....))).((((....))))..........))).....(.((((((((.((..........)).)))))))))...((((........))))........... ( -31.70) >DroEre_CAF1 45439 110 + 1 GAACCCGGCAG-A-GAGCCACCCAUCCUUGGGAUAGUAUAAACGGAUUAGAACCAUGUCUGGCAAAAAUCAACGCCAGACAUGGCAUUUCCUUAGCGAUAAGGAUCAG---CUCU ....(((((..-.-..))).((((....))))...........))....((.(((((((((((..........)))))))))))....((((((....))))))))..---.... ( -38.60) >DroYak_CAF1 46232 111 + 1 GAACCCGGCAA-AGGAGCCACCCAUCCUUGGGAUAGUAUAAACGGAUUAGGACCAUGUCUGGCAAAAAUCAACGCCAGACAUGGCAUUUCCUCACAGAUAAGGAUCAU---CACU ((.((.(((..-....))).((((....))))...........(((....(.(((((((((((..........))))))))))))...)))..........)).))..---.... ( -37.20) >consensus GAACCCGGGCA_AGGAGCCACCCAUCCUUGGGAUAGUAUAAACGGAUUAGGACCAUGUCUGGCAAAAAUCAACGCAAGACAUGGCAUUUCCUCACAGAUAAGGAUCAAC__CUCU ..............((.((.((((....))))...........(((......((((((((.((..........)).))))))))....)))..........)).))......... (-20.20 = -20.87 + 0.67)
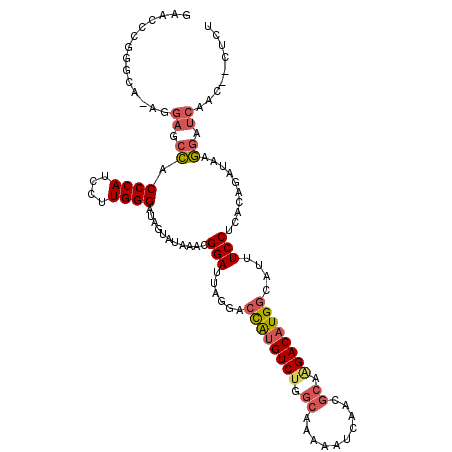
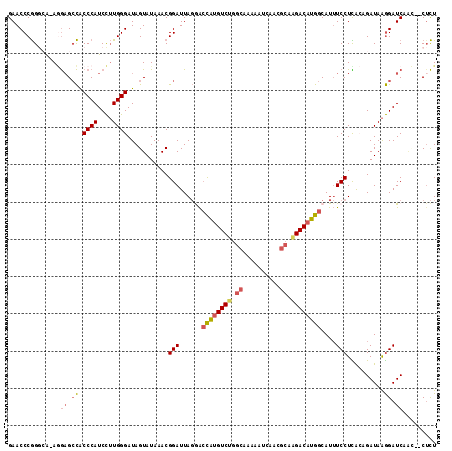
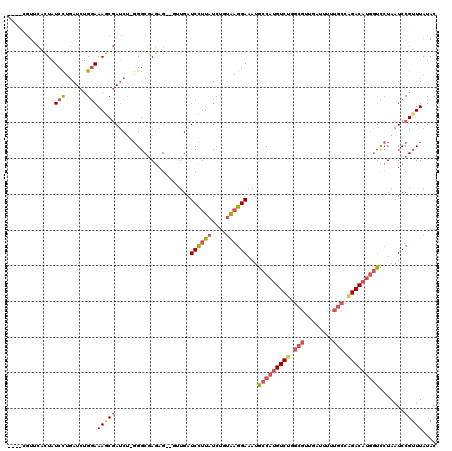
| Location | 13,345,341 – 13,345,455 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 83.31 |
| Mean single sequence MFE | -36.57 |
| Consensus MFE | -26.01 |
| Energy contribution | -26.93 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13345341 114 - 20766785 AGAGUUGUUGAUCCUUAUCUGUGAGGAAAUGCCAUGUCUUGCGUUGAUUUUUGCCAGACAUGGUCCUAAUCCGUUUAUACUAUCCCAAGGAUGGGUGGCUCCU-UGCCCGGGUUC ...........((((((....))))))...(((((((((.(((........))).)))))))))...((((((......((((((...))))))..(((....-.))))))))). ( -38.00) >DroPse_CAF1 42349 104 - 1 AAAG--AUUGAUCUCUAUCUCCAAGGAUAUGCCAUGUCUG---------UACGCAGGACUCAUCUGUAAUCCGUUUAUACUAUCCCAUAGAUGGGCACCGUUUCUGCCUGGGUUC ..((--((.(....).))))(((.((((((...)))))).---------...(((((((((((((((.....((....))......)))))))))......)))))).))).... ( -19.70) >DroSec_CAF1 44597 114 - 1 AGAGUUGUUGAUCCUUAUCUGUAAGGAAAUGCCAUGUCCUGCGUUGAUUUUUGCCAGACAUGGUCCUAAUCCGUUUAUACUAUCCCAAGGAUGGGUGGCUCCU-UGCCCGGGUUC ...........((((((....))))))...((((((((..(((........)))..))))))))...((((((......((((((...))))))..(((....-.))))))))). ( -35.70) >DroSim_CAF1 46583 114 - 1 AGAGUUGUUGAUCCUUAUCUGUGAGGAAAUGCCAUGUCUUGCGUUGAUUUUUGCCAGACAUGGUCCUAAUCCGUUUAUACUAUCCCAAGGAUGGGUGGCUCCU-CGCCCGGGUUC ...........((((((....))))))...(((((((((.(((........))).)))))))))...((((((......((((((...))))))..(((....-.))))))))). ( -37.40) >DroEre_CAF1 45439 110 - 1 AGAG---CUGAUCCUUAUCGCUAAGGAAAUGCCAUGUCUGGCGUUGAUUUUUGCCAGACAUGGUUCUAAUCCGUUUAUACUAUCCCAAGGAUGGGUGGCUC-U-CUGCCGGGUUC (((.---....((((((....))))))...(((((((((((((........))))))))))))))))(((((.......((((((...))))))..(((..-.-..)))))))). ( -44.30) >DroYak_CAF1 46232 111 - 1 AGUG---AUGAUCCUUAUCUGUGAGGAAAUGCCAUGUCUGGCGUUGAUUUUUGCCAGACAUGGUCCUAAUCCGUUUAUACUAUCCCAAGGAUGGGUGGCUCCU-UUGCCGGGUUC ....---....((((((....))))))...(((((((((((((........)))))))))))))...(((((.......((((((...))))))..(((....-..)))))))). ( -44.30) >consensus AGAG__GUUGAUCCUUAUCUGUAAGGAAAUGCCAUGUCUGGCGUUGAUUUUUGCCAGACAUGGUCCUAAUCCGUUUAUACUAUCCCAAGGAUGGGUGGCUCCU_UGCCCGGGUUC ...........((((((....))))))...(((((((((.(((........))).)))))))))...(((((.......(((((.....)))))..((.........))))))). (-26.01 = -26.93 + 0.92)
| Location | 13,345,375 – 13,345,494 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -17.31 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.678514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
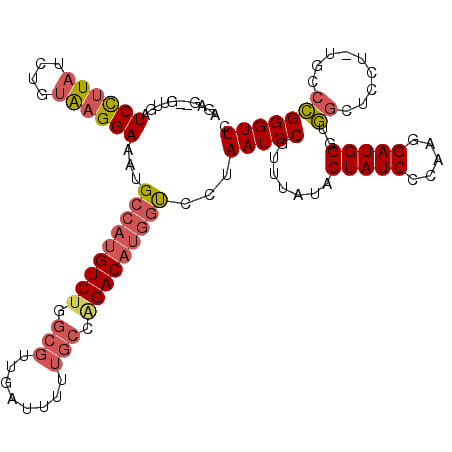
>2R_DroMel_CAF1 13345375 119 + 20766785 GUAUAAACGGAUUAGGACCAUGUCUGGCAAAAAUCAACGCAAGACAUGGCAUUUCCUCACAGAUAAGGAUCAACAACUCUCGCCC-AGAUCGCUUUCCAGAUCAGGAUAGUGAACGAAAC ........(((....(.((((((((.((..........)).)))))))))...)))((((.(((....)))............((-.((((........)))).))...))))....... ( -28.70) >DroPse_CAF1 42384 97 + 1 GUAUAAACGGAUUACAGAUGAGUCCUGCGUA---------CAGACAUGGCAUAUCCUUGGAGAUAGAGAUCAAU--CUUUUGGC--UGCUUGUUUUUCGGAUAAAGAGAA---------- ((((..(.(((((.(....)))))))..)))---------).((((.((((...((..((((((.(....).))--)))).)).--))))))))((((.......)))).---------- ( -17.40) >DroSec_CAF1 44631 116 + 1 GUAUAAACGGAUUAGGACCAUGUCUGGCAAAAAUCAACGCAGGACAUGGCAUUUCCUUACAGAUAAGGAUCAACAACUCUCGCCCAAGAUCGCUUUCCAGAUCAGGAUAGUGAACU---- ........((.....(.((((((((.((..........)).)))))))))...((((((....))))))..............)).((.(((((.(((......))).))))).))---- ( -30.30) >DroSim_CAF1 46617 116 + 1 GUAUAAACGGAUUAGGACCAUGUCUGGCAAAAAUCAACGCAAGACAUGGCAUUUCCUCACAGAUAAGGAUCAACAACUCUCGCCCAAGAUCGCUUUCCAGAUCAGGAUAGUGUACG---- ((((.....((((.((.((((((((.((..........)).))))))))....((((........))))..............))..))))(((.(((......))).))))))).---- ( -27.40) >DroEre_CAF1 45472 116 + 1 GUAUAAACGGAUUAGAACCAUGUCUGGCAAAAAUCAACGCCAGACAUGGCAUUUCCUUAGCGAUAAGGAUCAG---CUCUCGCCC-AGAUCGCUUUCCAGAUCAGGAUAGUGAACGAAAC ........((...(((.(((((((((((..........)))))))))))....((((((....))))))....---.)))...))-.(.(((((.(((......))).))))).)..... ( -38.20) >DroYak_CAF1 46266 116 + 1 GUAUAAACGGAUUAGGACCAUGUCUGGCAAAAAUCAACGCCAGACAUGGCAUUUCCUCACAGAUAAGGAUCAU---CACUCGCCC-AGAUCGAUUUCCAGAUCGGGAUAGUGCACGAAAC ........(((....(.(((((((((((..........))))))))))))...)))((...(((....)))..---((((..(((-.((((........)))))))..))))...))... ( -38.10) >consensus GUAUAAACGGAUUAGGACCAUGUCUGGCAAAAAUCAACGCAAGACAUGGCAUUUCCUCACAGAUAAGGAUCAAC__CUCUCGCCC_AGAUCGCUUUCCAGAUCAGGAUAGUGAACG____ ........(((......((((((((.((..........)).))))))))....))).....(((....)))...................((((.(((......))).))))........ (-17.31 = -18.07 + 0.75)

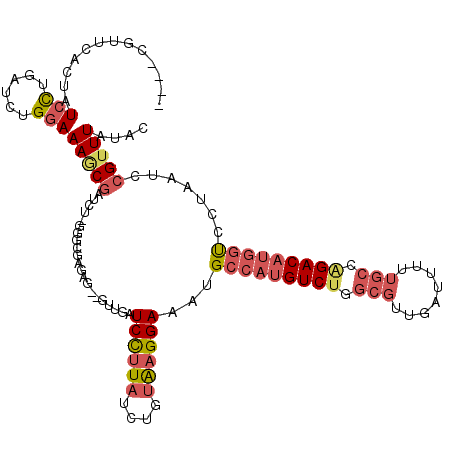
| Location | 13,345,375 – 13,345,494 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -20.89 |
| Energy contribution | -22.17 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
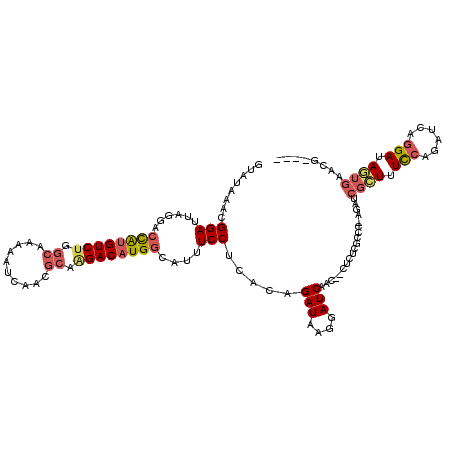
>2R_DroMel_CAF1 13345375 119 - 20766785 GUUUCGUUCACUAUCCUGAUCUGGAAAGCGAUCU-GGGCGAGAGUUGUUGAUCCUUAUCUGUGAGGAAAUGCCAUGUCUUGCGUUGAUUUUUGCCAGACAUGGUCCUAAUCCGUUUAUAC (((((.(((((..(((......)))(((.((((.-.(((....)))...)))))))....))))))))))(((((((((.(((........))).)))))))))................ ( -35.90) >DroPse_CAF1 42384 97 - 1 ----------UUCUCUUUAUCCGAAAAACAAGCA--GCCAAAAG--AUUGAUCUCUAUCUCCAAGGAUAUGCCAUGUCUG---------UACGCAGGACUCAUCUGUAAUCCGUUUAUAC ----------...............((((..(((--(.((..((--((.(....).))))....((.....)).)).)))---------)..(((((.....))))).....)))).... ( -11.50) >DroSec_CAF1 44631 116 - 1 ----AGUUCACUAUCCUGAUCUGGAAAGCGAUCUUGGGCGAGAGUUGUUGAUCCUUAUCUGUAAGGAAAUGCCAUGUCCUGCGUUGAUUUUUGCCAGACAUGGUCCUAAUCCGUUUAUAC ----((.((.((.(((......))).)).)).))((((((.((........((((((....))))))...((((((((..(((........)))..)))))))).....))))))))... ( -31.50) >DroSim_CAF1 46617 116 - 1 ----CGUACACUAUCCUGAUCUGGAAAGCGAUCUUGGGCGAGAGUUGUUGAUCCUUAUCUGUGAGGAAAUGCCAUGUCUUGCGUUGAUUUUUGCCAGACAUGGUCCUAAUCCGUUUAUAC ----....(((..(((......)))(((.((((...(((....)))...)))))))....))).(((...(((((((((.(((........))).))))))))).....)))........ ( -33.80) >DroEre_CAF1 45472 116 - 1 GUUUCGUUCACUAUCCUGAUCUGGAAAGCGAUCU-GGGCGAGAG---CUGAUCCUUAUCGCUAAGGAAAUGCCAUGUCUGGCGUUGAUUUUUGCCAGACAUGGUUCUAAUCCGUUUAUAC .............(((......))).((((((..-((((....)---)....))..))))))..(((...(((((((((((((........))))))))))))).....)))........ ( -42.70) >DroYak_CAF1 46266 116 - 1 GUUUCGUGCACUAUCCCGAUCUGGAAAUCGAUCU-GGGCGAGUG---AUGAUCCUUAUCUGUGAGGAAAUGCCAUGUCUGGCGUUGAUUUUUGCCAGACAUGGUCCUAAUCCGUUUAUAC ...((((.((((..(((((((........)))).-)))..))))---))))((((((....))))))...(((((((((((((........)))))))))))))................ ( -44.50) >consensus ____CGUUCACUAUCCUGAUCUGGAAAGCGAUCU_GGGCGAGAG__GUUGAUCCUUAUCUGUAAGGAAAUGCCAUGUCUGGCGUUGAUUUUUGCCAGACAUGGUCCUAAUCCGUUUAUAC .............(((......)))(((((.....................((((((....))))))...(((((((((.(((........))).))))))))).......))))).... (-20.89 = -22.17 + 1.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:22 2006