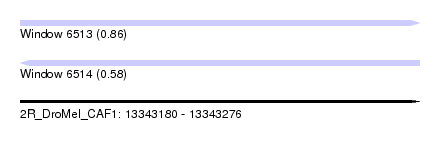
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,343,180 – 13,343,276 |
| Length | 96 |
| Max. P | 0.855834 |

| Location | 13,343,180 – 13,343,276 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 77.81 |
| Mean single sequence MFE | -26.83 |
| Consensus MFE | -19.92 |
| Energy contribution | -19.73 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13343180 96 + 20766785 --------------UCUCUA-CAAACAGUCCUUUGAUUAUACGGAAUUGUGCCAGCCGGGCACGCUGAUCGAUGCCAAUGGCAGCAUACCCGUCAGCGUGAACAGCAUCCA- --------------......-((((......)))).......(((...(((((.....)))))((((.((.((((..((((........))))..)))))).)))).))).- ( -27.60) >DroPse_CAF1 39679 94 + 1 ---------------CCAUCU--UCCAGUCCUUCGAUUACACGGAACUGUGCCAGCCGGGCACCCUCAUCGAUGCCAAUGGCAGCAUACCCGUGUCCGUGAACAGCAUCCA- ---------------......--....((..((((...((((((...(((((..(((.((((.(......).))))...))).))))).))))))...))))..)).....- ( -27.70) >DroWil_CAF1 54436 98 + 1 U-------------GCCUCU-UCUACAGUCAUUCGACUACACGGAAUUAUGCCAGCCGGGCACCUUGAUCGAUGCCAAUGGCAGCAUUCCCGUUUCGGUCAAUAGCCUUCAA .-------------(((...-.....((((....))))....(((((..(((((....((((.(......).))))..)))))..)))))......)))............. ( -25.00) >DroMoj_CAF1 50247 109 + 1 -CAUAUUUGCAACUUCUUUU-CGAACAGUCCUUCGACUAUACGGAACUGUGUCAGCCGGGCACCCUCAUCGAUGCCAAUGGCAGCAUACCCGUUUCAGUGAAUAGCAUACA- -..((((..(.........(-((((......)))))....((((...(((((..(((.((((.(......).))))...))).))))).))))....)..)))).......- ( -24.90) >DroAna_CAF1 42837 91 + 1 --------------UU------AUUUAGUCCUUCGAUUAUACGGAACUGUGCCAGCCGGGCACGCUGAUCGAUGCCAAUGGCAGCAUACCCGUCAGUGUGAACAGUAUCCA- --------------..------...(((((....)))))...(((((((((((.....)))((((((((..((((........))))....))))))))..))))).))).- ( -28.10) >DroPer_CAF1 39947 94 + 1 ---------------CCAUCU--UCCAGUCCUUCGAUUACACGGAACUGUGCCAGCCGGGCACCCUCAUCGAUGCCAAUGGCAGCAUACCCGUGUCCGUGAACAGCAUCCA- ---------------......--....((..((((...((((((...(((((..(((.((((.(......).))))...))).))))).))))))...))))..)).....- ( -27.70) >consensus ______________UCCAUC___UACAGUCCUUCGAUUACACGGAACUGUGCCAGCCGGGCACCCUCAUCGAUGCCAAUGGCAGCAUACCCGUCUCCGUGAACAGCAUCCA_ ...........................((..((((.....((((...(((((..(((.(((((.......).))))...))).))))).)))).....))))..))...... (-19.92 = -19.73 + -0.19)


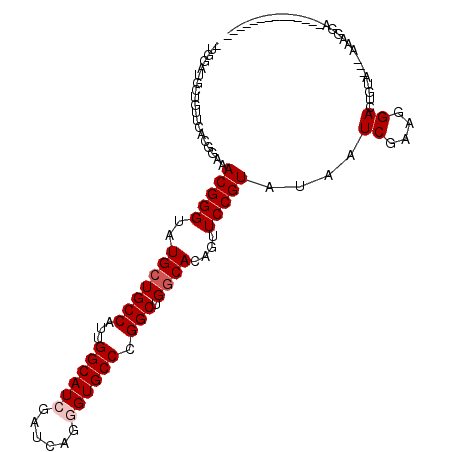
| Location | 13,343,180 – 13,343,276 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.81 |
| Mean single sequence MFE | -32.28 |
| Consensus MFE | -21.08 |
| Energy contribution | -21.58 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13343180 96 - 20766785 -UGGAUGCUGUUCACGCUGACGGGUAUGCUGCCAUUGGCAUCGAUCAGCGUGCCCGGCUGGCACAAUUCCGUAUAAUCAAAGGACUGUUUG-UAGAGA-------------- -.....((((..((((((((((...(((((......))))))).))))))))..))))....((((.(((...........)))....)))-).....-------------- ( -28.90) >DroPse_CAF1 39679 94 - 1 -UGGAUGCUGUUCACGGACACGGGUAUGCUGCCAUUGGCAUCGAUGAGGGUGCCCGGCUGGCACAGUUCCGUGUAAUCGAAGGACUGGA--AGAUGG--------------- -.......(.(((.((((((((((..((.(((((..((((((......))))))....)))))))..)))))))..((....)))))))--).)...--------------- ( -32.80) >DroWil_CAF1 54436 98 - 1 UUGAAGGCUAUUGACCGAAACGGGAAUGCUGCCAUUGGCAUCGAUCAAGGUGCCCGGCUGGCAUAAUUCCGUGUAGUCGAAUGACUGUAGA-AGAGGC-------------A ......(((.(((((....((((((((((((((...((((((......)))))).))).)))))..))))))...)))))....((.....-)).)))-------------. ( -33.90) >DroMoj_CAF1 50247 109 - 1 -UGUAUGCUAUUCACUGAAACGGGUAUGCUGCCAUUGGCAUCGAUGAGGGUGCCCGGCUGACACAGUUCCGUAUAGUCGAAGGACUGUUCG-AAAAGAAGUUGCAAAUAUG- -(((.(((..(((((((...(((((((.((..(((((....))))))).))))))).......))))..((.((((((....)))))).))-....)))...))).)))..- ( -34.10) >DroAna_CAF1 42837 91 - 1 -UGGAUACUGUUCACACUGACGGGUAUGCUGCCAUUGGCAUCGAUCAGCGUGCCCGGCUGGCACAGUUCCGUAUAAUCGAAGGACUAAAU------AA-------------- -.(((.(((((.(.((....(((((((((((..((((....)))))))))))))))..))).))))))))......((....))......------..-------------- ( -31.20) >DroPer_CAF1 39947 94 - 1 -UGGAUGCUGUUCACGGACACGGGUAUGCUGCCAUUGGCAUCGAUGAGGGUGCCCGGCUGGCACAGUUCCGUGUAAUCGAAGGACUGGA--AGAUGG--------------- -.......(.(((.((((((((((..((.(((((..((((((......))))))....)))))))..)))))))..((....)))))))--).)...--------------- ( -32.80) >consensus _UGGAUGCUGUUCACGGAAACGGGUAUGCUGCCAUUGGCAUCGAUCAGGGUGCCCGGCUGGCACAGUUCCGUAUAAUCGAAGGACUGUA___AAAGGA______________ ...................(((((..(((((((...((((((......)))))).))).))))....)))))....((....))............................ (-21.08 = -21.58 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:18 2006