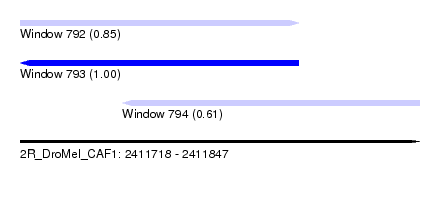
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,411,718 – 2,411,847 |
| Length | 129 |
| Max. P | 0.998030 |

| Location | 2,411,718 – 2,411,808 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 85.49 |
| Mean single sequence MFE | -23.51 |
| Consensus MFE | -21.08 |
| Energy contribution | -21.09 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2411718 90 + 20766785 GCUGCCAGCGAGUGCCUGACCUUUUUGCCAGCCAUGCAGUAGCUCC----CACCGCCUUCAGCUUUCAGCUUUCAGCUUUCGCCUUGCUGGCUU ...(((((((((.((..((.((..((((.......)))).)).)).----....((....(((.....)))....))....))))))))))).. ( -25.70) >DroSec_CAF1 68114 76 + 1 GCUGCCAGCGAGUGCCUGACCUUUUUGCCAGCCAUGCAGUAGCUCC----CACCAC--------------UCUCAGCUUUCGCCUUGCUGGCUU ...(((((((((((...((.((..((((.......)))).)).)).----...)))--------------))...((....))...)))))).. ( -22.10) >DroSim_CAF1 25222 76 + 1 GCUGCCAGCGAGUGCCUGACCUUUUUGCCAGCCAUGCAGUAGCUCC----CACCAC--------------UCUCAGCUUUCGCCUUGCUGGCUU ...(((((((((((...((.((..((((.......)))).)).)).----...)))--------------))...((....))...)))))).. ( -22.10) >DroYak_CAF1 24520 80 + 1 GCUGCCAGCGAGUGCCUAACCUUUUUGCCAGCCAUGCAGUAGCUGCCUUUCACUGC--------------CUUCAGCUUUCGCCUUGCUGGCUU ...(((((((((.((.........((((.......)))).(((((...........--------------...)))))...))))))))))).. ( -24.14) >consensus GCUGCCAGCGAGUGCCUGACCUUUUUGCCAGCCAUGCAGUAGCUCC____CACCAC______________UCUCAGCUUUCGCCUUGCUGGCUU ...(((((((((.((.........((((.......)))).((((..............................))))...))))))))))).. (-21.08 = -21.09 + 0.00)


| Location | 2,411,718 – 2,411,808 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 85.49 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -33.00 |
| Energy contribution | -32.88 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.57 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2411718 90 - 20766785 AAGCCAGCAAGGCGAAAGCUGAAAGCUGAAAGCUGAAGGCGGUG----GGAGCUACUGCAUGGCUGGCAAAAAGGUCAGGCACUCGCUGGCAGC ..((((((..(((....)))...(((.....)))((..((((((----(...))))))).((.(((((......))))).)).))))))))... ( -37.70) >DroSec_CAF1 68114 76 - 1 AAGCCAGCAAGGCGAAAGCUGAGA--------------GUGGUG----GGAGCUACUGCAUGGCUGGCAAAAAGGUCAGGCACUCGCUGGCAGC ..((((((..(((....)))(((.--------------(..(((----(...))))..).((.(((((......))))).)))))))))))... ( -37.00) >DroSim_CAF1 25222 76 - 1 AAGCCAGCAAGGCGAAAGCUGAGA--------------GUGGUG----GGAGCUACUGCAUGGCUGGCAAAAAGGUCAGGCACUCGCUGGCAGC ..((((((..(((....)))(((.--------------(..(((----(...))))..).((.(((((......))))).)))))))))))... ( -37.00) >DroYak_CAF1 24520 80 - 1 AAGCCAGCAAGGCGAAAGCUGAAG--------------GCAGUGAAAGGCAGCUACUGCAUGGCUGGCAAAAAGGUUAGGCACUCGCUGGCAGC ..((((((..(((....)))((..--------------((((((.........)))))).((.(((((......))))).)).))))))))... ( -32.30) >consensus AAGCCAGCAAGGCGAAAGCUGAAA______________GCGGUG____GGAGCUACUGCAUGGCUGGCAAAAAGGUCAGGCACUCGCUGGCAGC ..((((((..(((....)))(((...............((((((.........)))))).((.(((((......))))).)))))))))))... (-33.00 = -32.88 + -0.12)


| Location | 2,411,751 – 2,411,847 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 85.90 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -16.09 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612072 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2411751 96 - 20766785 AAGGGGAUCCAAUAAAUUCGAUUGCAACCGAAGCAUGAAAAGCCAGCAAGGCGAAAGCUGAAAGCUGAAAGCUGAAGGCGGUG----GGAGCUACUGCAU ..((....))......((((..(((.......))))))).(((((((..(((....)))....))))...)))....((((((----(...))))))).. ( -24.50) >DroSec_CAF1 68147 82 - 1 AAGGGGAUUCAAUAAAUUCGAUUGCAAUCGAAGCAUGAAAAGCCAGCAAGGCGAAAGCUGAGA--------------GUGGUG----GGAGCUACUGCAU ....((.((((.....((((((....))))))...))))...)).((..(((....)))...(--------------(((((.----...)))))))).. ( -22.10) >DroSim_CAF1 25255 82 - 1 AAGGGGAUCCAAUAAAUUCGAUUGCAACCGAAGCAUGAAAAGCCAGCAAGGCGAAAGCUGAGA--------------GUGGUG----GGAGCUACUGCAU .((..(.(((......((((..(((.......)))))))..((((.(..(((....)))..).--------------.)))).----))).)..)).... ( -19.40) >DroYak_CAF1 24553 86 - 1 AAGGGGAUCCAAUAAAUUCGAUUGCAACCGAAGCAUGAAAAGCCAGCAAGGCGAAAGCUGAAG--------------GCAGUGAAAGGCAGCUACUGCAU ..((....))......(((.(((((.......((.((......))))..(((....)))....--------------))))))))..((((...)))).. ( -18.60) >consensus AAGGGGAUCCAAUAAAUUCGAUUGCAACCGAAGCAUGAAAAGCCAGCAAGGCGAAAGCUGAAA______________GCGGUG____GGAGCUACUGCAU ....((..........((((........))))..........)).(((.(((....)))..................(((((........)))))))).. (-16.09 = -16.40 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:46 2006