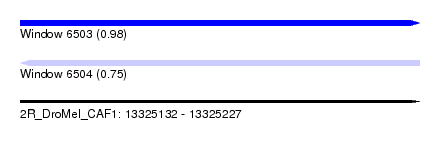
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,325,132 – 13,325,227 |
| Length | 95 |
| Max. P | 0.981394 |

| Location | 13,325,132 – 13,325,227 |
|---|---|
| Length | 95 |
| Sequences | 5 |
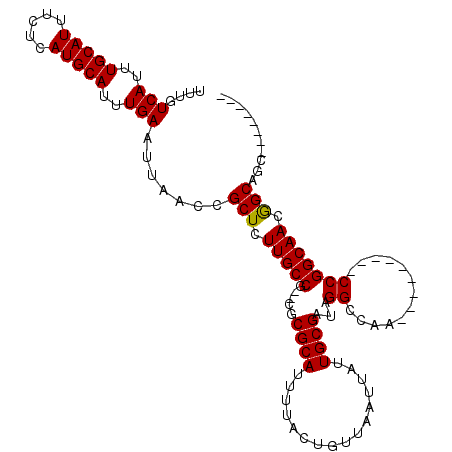
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.24 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -23.21 |
| Energy contribution | -22.97 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
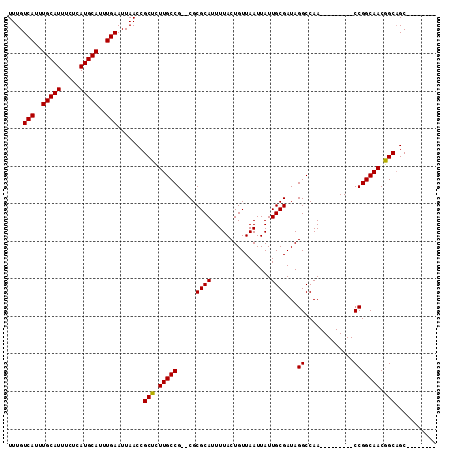
>2R_DroMel_CAF1 13325132 95 + 20766785 UUUGUCAUUUGCAUUUCUCAUGCAUUUGAAUUAACCGCUCUUGCCG--CGCGCAUUUUACUGUUAAUUAUUGCGAUAGGCCAA---------CCGGCAACGGCAGC-------- ....(((..(((((.....)))))..))).......(((.(((((.--..((((................))))...((....---------))))))).)))...-------- ( -24.49) >DroPse_CAF1 22661 106 + 1 UUUGUCAUUUGCAUUUCUCAUGCAUUUGAAUUAACCGCUUUUGCCGGGCGCGCAUUUUACUGUUAAUUAUUGCGAUAGGCCAACAUCGUCAACCGGCAACAGCAGC-------- ....(((..(((((.....)))))..))).......(((.(((((((...((((................))))...(((.......)))..))))))).)))...-------- ( -30.19) >DroSim_CAF1 25850 95 + 1 UUUGUCAUUUGCAUUUCUCAUGCAUUUGAAUUAACCGCUCUUGCCG--CGCGCAUUUUACUGUUAAUUAUUGCGAUAGGCCAA---------CCGGCAACGGCAGC-------- ....(((..(((((.....)))))..))).......(((.(((((.--..((((................))))...((....---------))))))).)))...-------- ( -24.49) >DroEre_CAF1 25251 103 + 1 UUUGUCAUUUGCAUUUCUCAUGCAUUUGAAUUAACCGCUCUUGCCG--CGCGCAUUUUACUGUUAAUUAUUGCGAUAGGCCAA---------CCGGCAACGGCUGCAACAUUGC ....(((..(((((.....)))))..))).......((....(((.--..((((................))))...)))...---------(((....)))..))........ ( -25.19) >DroPer_CAF1 22793 106 + 1 UUUGUCAUUUGCAUUUCUCAUGCAUUUGAAUUAACCGCUUUUGCCGGGCGCGCAUUUUACUGUUAAUUAUUGCGAUAGGCCAACAUCGUCAACCGGCAACAGCAGC-------- ....(((..(((((.....)))))..))).......(((.(((((((...((((................))))...(((.......)))..))))))).)))...-------- ( -30.19) >consensus UUUGUCAUUUGCAUUUCUCAUGCAUUUGAAUUAACCGCUCUUGCCG__CGCGCAUUUUACUGUUAAUUAUUGCGAUAGGCCAA_________CCGGCAACGGCAGC________ ....(((..(((((.....)))))..))).......(((.(((((.....((((................))))...((.............))))))).)))........... (-23.21 = -22.97 + -0.24)

| Location | 13,325,132 – 13,325,227 |
|---|---|
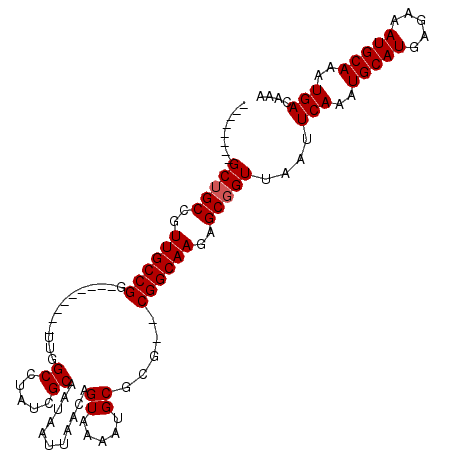
| Length | 95 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.24 |
| Mean single sequence MFE | -27.39 |
| Consensus MFE | -21.64 |
| Energy contribution | -21.84 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13325132 95 - 20766785 --------GCUGCCGUUGCCGG---------UUGGCCUAUCGCAAUAAUUAACAGUAAAAUGCGCG--CGGCAAGAGCGGUUAAUUCAAAUGCAUGAGAAAUGCAAAUGACAAA --------(((((..(((((((---------....))...((((................))))..--.)))))..)))))....(((..(((((.....)))))..))).... ( -24.39) >DroPse_CAF1 22661 106 - 1 --------GCUGCUGUUGCCGGUUGACGAUGUUGGCCUAUCGCAAUAAUUAACAGUAAAAUGCGCGCCCGGCAAAAGCGGUUAAUUCAAAUGCAUGAGAAAUGCAAAUGACAAA --------((((((.((((((((((.(((((......)))))))).........((.....))....))))))).))))))....(((..(((((.....)))))..))).... ( -31.50) >DroSim_CAF1 25850 95 - 1 --------GCUGCCGUUGCCGG---------UUGGCCUAUCGCAAUAAUUAACAGUAAAAUGCGCG--CGGCAAGAGCGGUUAAUUCAAAUGCAUGAGAAAUGCAAAUGACAAA --------(((((..(((((((---------....))...((((................))))..--.)))))..)))))....(((..(((((.....)))))..))).... ( -24.39) >DroEre_CAF1 25251 103 - 1 GCAAUGUUGCAGCCGUUGCCGG---------UUGGCCUAUCGCAAUAAUUAACAGUAAAAUGCGCG--CGGCAAGAGCGGUUAAUUCAAAUGCAUGAGAAAUGCAAAUGACAAA (((....)))((((((((((((---------....))...((((................))))..--.))))...))))))...(((..(((((.....)))))..))).... ( -25.19) >DroPer_CAF1 22793 106 - 1 --------GCUGCUGUUGCCGGUUGACGAUGUUGGCCUAUCGCAAUAAUUAACAGUAAAAUGCGCGCCCGGCAAAAGCGGUUAAUUCAAAUGCAUGAGAAAUGCAAAUGACAAA --------((((((.((((((((((.(((((......)))))))).........((.....))....))))))).))))))....(((..(((((.....)))))..))).... ( -31.50) >consensus ________GCUGCCGUUGCCGG_________UUGGCCUAUCGCAAUAAUUAACAGUAAAAUGCGCG__CGGCAAGAGCGGUUAAUUCAAAUGCAUGAGAAAUGCAAAUGACAAA ........(((((..((((((.............((.....))...........((.....)).....))))))..)))))....(((..(((((.....)))))..))).... (-21.64 = -21.84 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:08 2006