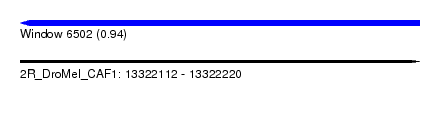
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,322,112 – 13,322,220 |
| Length | 108 |
| Max. P | 0.941697 |

| Location | 13,322,112 – 13,322,220 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -12.15 |
| Energy contribution | -12.43 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
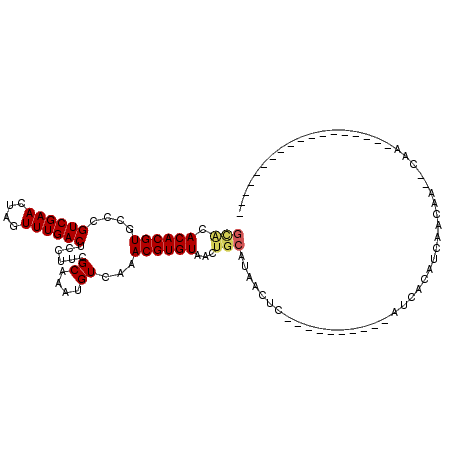
>2R_DroMel_CAF1 13322112 108 - 20766785 GCACACACGUGGCCGUCGAACUAGUUUGACUCCUUCGCAAAUGUCAAACGUGUAACUGCAUAACUCAGUGCGGUUCAUCACAUCGGCAAAACAACAAAACAACC----AAAC .(((....)))((((........(((((((............)))))))(((.((((((((......))))))))...)))..)))).................----.... ( -28.70) >DroVir_CAF1 22830 82 - 1 GCACACACGUGCCCGUCGAACUAGUUUGACUCCUUCGCAAAUGUCAAACGUGUAACUGCAUAACUC----------AUCACAUCAACAA--CAA------------------ ((((....))))..((.((....(((((((............)))))))((((....)))).....----------......)).))..--...------------------ ( -15.20) >DroGri_CAF1 22013 82 - 1 GCACACACGUGCCCGUCGAACUAGUUUGACUCCUUCGCAAAUGUCAAACGUGUAACUGCAUAACUC----------AUCACAUCAACAA--CAA------------------ ((((....))))..((.((....(((((((............)))))))((((....)))).....----------......)).))..--...------------------ ( -15.20) >DroEre_CAF1 22069 100 - 1 GCACACACGUGGCCGUCGAACUAGUUUGACUCCUUCGCAAAUGUCAAACGUGUAACUGCAUAACUCAGUGCGGUUCAUCACAUCAG--------CAAAAC----AACAAAAC .(((....)))...((.((....(((((((............)))))))(((.((((((((......))))))))...))).)).)--------).....----........ ( -23.60) >DroMoj_CAF1 23244 82 - 1 GUGCACACGUGCCCGUCGAACUAGUUUGACUCCUUCGCAAAUGUCAAACGUGUAACUGCAUAACUC----------AUCACAUCAACAA--CAA------------------ ((((((((((....((((((....))))))......((....))...))))))....)))).....----------.............--...------------------ ( -14.20) >DroAna_CAF1 21839 103 - 1 ---CACACGUGCCCGUCGAACUAGUUUGACUCCUUCGCAAAUGUCAAACGUGUAACUGCAUAACUCAACGCGGUUCAUCACAUCAACAAAACA-CUAAACAACA----ACA- ---.....(((...((.((....(((((((............)))))))(((.((((((..........))))))...))).)).))....))-).........----...- ( -19.10) >consensus GCACACACGUGCCCGUCGAACUAGUUUGACUCCUUCGCAAAUGUCAAACGUGUAACUGCAUAACUC__________AUCACAUCAACAA__CAA__________________ (((.((((((....((((((....))))))......((....))...))))))...)))..................................................... (-12.15 = -12.43 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:06 2006