| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,307,129 – 13,307,245 |
| Length | 116 |
| Max. P | 0.850700 |

| Location | 13,307,129 – 13,307,245 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.39 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -25.74 |
| Energy contribution | -25.46 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
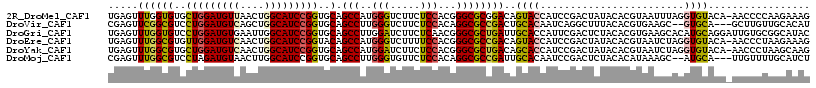
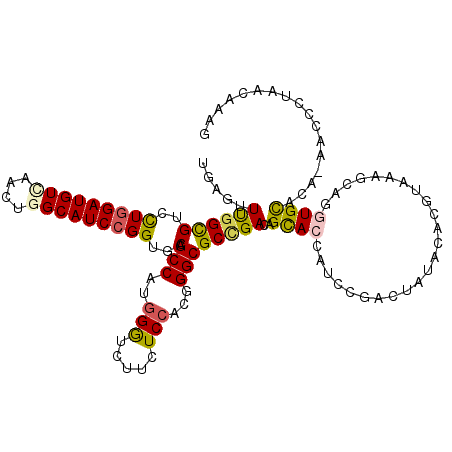
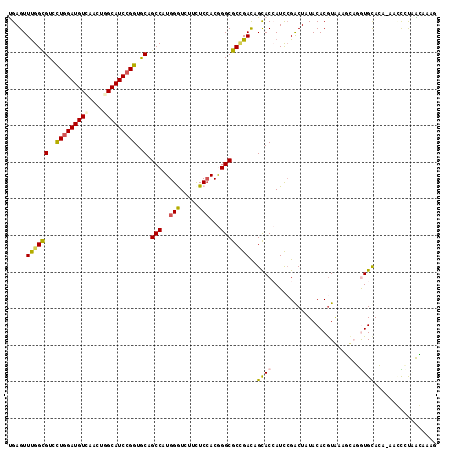
>2R_DroMel_CAF1 13307129 116 + 20766785 UGAGUUUGGUGUGCUGGAUGUUAACUGGCAUCCGGUGCAGCCAUGGGUCUUCUCCACGGGCGCGGACAGUACCAUCCGACUAUACACGUAAUUUAGGUGUACA-AACCCCAAGAAAG ...(((((.((..(((((((((....)))))))))..))(((...((......))...))).((((........))))....(((((.((...)).)))))))-))).......... ( -37.60) >DroVir_CAF1 5700 112 + 1 CGAGUUCGGCGUCCUGGAUGUCAGCUGGCAUCCGGUGCAGCCUUGGGUCUUCUCCACAGGCGCCGACUGCACAAUCAGGCUUUACACGUGAAGC--GUGCA---GCUUGUUGCACAU .....((((((..(((((((((....)))))))))..).((((((((.....)))).)))))))))(((......)))((((((....))))))--(((((---(....)))))).. ( -50.70) >DroGri_CAF1 6292 117 + 1 UGAGUUUGGUGUCCUGGAUGUGAAUUGGCAUCCGGUGCAGCCUUGGAUCUUCUCAACGGGCGCUGAUUGCACCAUUCGACUCUACACGUGAAGCACAUGCAGGAUUGUGCGGCAUAC .(((((((((((.((((((((......))))))))..((((((((...........)))).))))...))))))...))))).....(((..(((((........)))))..))).. ( -41.10) >DroEre_CAF1 7468 116 + 1 UGAGUUUGGCGUGUUGGAUGUCAACUGGCAUCCGGUACAGCCAUGGGUCUUUUCCACGGGCGCCGACAGUACCAUCCGACUAUACACGUAAUCUAGGUGUACA-AACCCUAAGAAAG ........((((((.(((((((....)))))))(((((.(((.((((.....))))..)))(....).)))))..........))))))..((((((.((...-.))))).)))... ( -37.00) >DroYak_CAF1 7435 116 + 1 UGAGUUUGGCGUGCUGGAUGUCAACUGGCAUCCGGUGCAGCCAUGGAUCUUCUCCACGGGCGCUGACAGCACCAUCCGACUAUACACGUAAUCUAGGUGUACA-AACCCUAAGCAAG ...((((((((..(((((((((....)))))))))..).(((.((((.....))))..))))).....(((((....((.(((....))).))..))))).))-))).......... ( -40.20) >DroMoj_CAF1 5764 112 + 1 CGAGUUUGGCGUCCUAGAUGUAACUUGGCAUCCGGUGCAGCCUUGGGUGUUCUCCACAGGCGCCGAUUGCACAAUCCGACUCUACACAUAAAGC--AUGCA---UUGUUUUGCAUCU .(((((.((((..(..(((((......)))))..)..).((((((((.....)))).)))))))((((....)))).))))).........((.--(((((---......))))))) ( -34.70) >consensus UGAGUUUGGCGUCCUGGAUGUCAACUGGCAUCCGGUGCAGCCAUGGGUCUUCUCCACGGGCGCCGACAGCACCAUCCGACUAUACACGUAAAGCAGGUGCACA_AACCCUAACAAAG .....((((((..(((((((((....)))))))))..).(((..(((.....)))...))))))))..((((........................))))................. (-25.74 = -25.46 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:01 2006