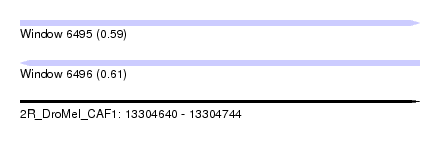
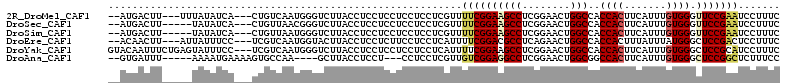
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,304,640 – 13,304,744 |
| Length | 104 |
| Max. P | 0.610733 |

| Location | 13,304,640 – 13,304,744 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -19.55 |
| Energy contribution | -19.67 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588066 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
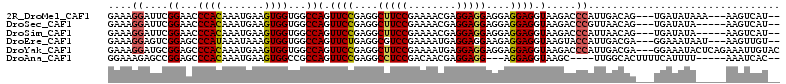
>2R_DroMel_CAF1 13304640 104 + 20766785 GAAAGGAUUCGGAACCCACAAAUGAAGUGGUGGCCAGUUCCGAGGCUUCCGAAAACGAGGAGGAGGAGGAGGUAAGACCCAUUGACAG---UGAUAUAAA---AAGUCAU-- ....((.((((((((((((..........))))...))))))))((((((.....(.....).....)))))).....))..((((..---.........---..)))).-- ( -25.62) >DroSec_CAF1 4940 102 + 1 GAAAGGAUUCGGAACCCACAAAUGAAGUGGUGGCCAGUUCCGAGGCUUCCGAAAACGAGGAGGAGGAGGAGGUAAGACCCGUUAACAG---UGAUAUA-----AAGUCAU-- ....((.((((((((((((..........))))...))))))))((((((.....(.....).....)))))).....)).......(---((((...-----..)))))-- ( -25.90) >DroSim_CAF1 4066 102 + 1 GAAAGGAUUCGGAACCCACAAAUGAAGUGGUGGCCAGUUCCGAGGCUUCCGAAAACGAGGAGGAGGAGGAGGUAAGACCCAUUAACAG---UGAUAUA-----AAGUCAU-- ....((.((((((((((((..........))))...))))))))((((((.....(.....).....)))))).....)).......(---((((...-----..)))))-- ( -25.90) >DroEre_CAF1 4973 104 + 1 GAAAGGAGUCGGAGCCCAUAAAUAAAGUGGUGGCCAGUUCUGAGGCGUCCGAAAAUGAGGAGGAAGAGGAGGUAAGUACCAUUGACGA---GGAAAUAAU---AAGUUGU-- .......(((((...))........((((((((((..((((....(.(((........))).).))))..)))...))))))))))..---.........---.......-- ( -21.20) >DroYak_CAF1 4943 109 + 1 GAAAGGAUGCGGAGCCCACAAAUGAAGUGGUGGCCAGUUCCGAGGCUUCCGAAAAUGAGGAGGAGGAGGAGGUAAGACCCAUUGACGA---GGAAAUACUCAGAAAUUGUAC .....((((.((..(((((.......)))).)(((..((((....(((((........))))).))))..)))....))))))...((---(......)))........... ( -27.90) >DroAna_CAF1 3138 98 + 1 GGAAAGAGCCGGAGCCCACAAAUGAAGUGGCCGCCAGUUCCGAGGCCUCCGACAACGAGGAGG---AGGAGGUAAGC----UUGGCACUUUUCAUUUU-----AAAUCAC-- ((((((.((((.(((((((.......))))..(((...(((....(((((........)))))---.))))))..))----))))).)))))).....-----.......-- ( -34.30) >consensus GAAAGGAUUCGGAACCCACAAAUGAAGUGGUGGCCAGUUCCGAGGCUUCCGAAAACGAGGAGGAGGAGGAGGUAAGACCCAUUGACAG___UGAUAUA_____AAGUCAU__ ....((....((...((((.......))))...))(.((((....(((((........)))))....)))).).....))................................ (-19.55 = -19.67 + 0.11)
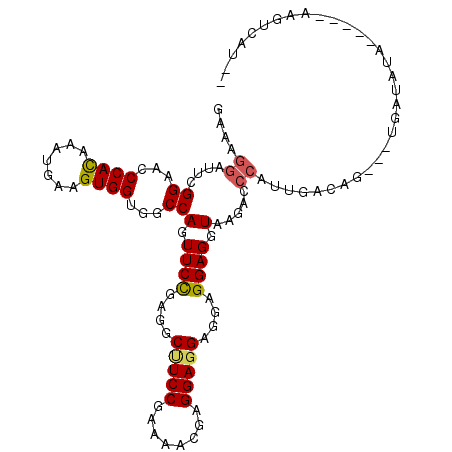
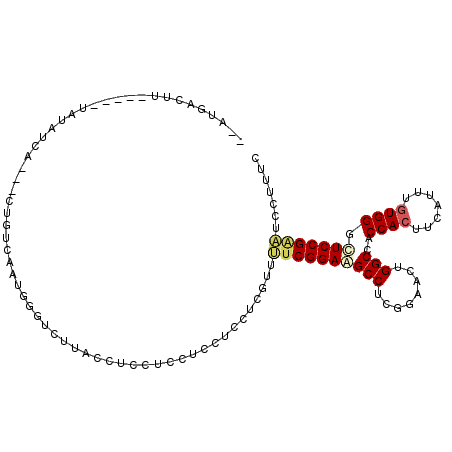
| Location | 13,304,640 – 13,304,744 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 81.62 |
| Mean single sequence MFE | -21.91 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.65 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13304640 104 - 20766785 --AUGACUU---UUUAUAUCA---CUGUCAAUGGGUCUUACCUCCUCCUCCUCCUCGUUUUCGGAAGCCUCGGAACUGGCCACCACUUCAUUUGUGGGUUCCGAAUCCUUUC --.((((..---.........---..))))..((......))..............(..((((((((((........)))..((((.......)))).)))))))..).... ( -23.52) >DroSec_CAF1 4940 102 - 1 --AUGACUU-----UAUAUCA---CUGUUAACGGGUCUUACCUCCUCCUCCUCCUCGUUUUCGGAAGCCUCGGAACUGGCCACCACUUCAUUUGUGGGUUCCGAAUCCUUUC --.((((..-----.......---..))))..((......))..............(..((((((((((........)))..((((.......)))).)))))))..).... ( -21.80) >DroSim_CAF1 4066 102 - 1 --AUGACUU-----UAUAUCA---CUGUUAAUGGGUCUUACCUCCUCCUCCUCCUCGUUUUCGGAAGCCUCGGAACUGGCCACCACUUCAUUUGUGGGUUCCGAAUCCUUUC --..(((((-----.......---........)))))...................(..((((((((((........)))..((((.......)))).)))))))..).... ( -21.76) >DroEre_CAF1 4973 104 - 1 --ACAACUU---AUUAUUUCC---UCGUCAAUGGUACUUACCUCCUCUUCCUCCUCAUUUUCGGACGCCUCAGAACUGGCCACCACUUUAUUUAUGGGCUCCGACUCCUUUC --.......---.........---..(((...(((....)))....................(((.(((.(((((.(((...))).))).....)))))))))))....... ( -14.60) >DroYak_CAF1 4943 109 - 1 GUACAAUUUCUGAGUAUUUCC---UCGUCAAUGGGUCUUACCUCCUCCUCCUCCUCAUUUUCGGAAGCCUCGGAACUGGCCACCACUUCAUUUGUGGGCUCCGCAUCCUUUC ((.....(((((((..(((((---..(..((((((..................))))))..)))))).)))))))..((...((((.......))))...))))........ ( -20.97) >DroAna_CAF1 3138 98 - 1 --GUGAUUU-----AAAAUGAAAAGUGCCAA----GCUUACCUCCU---CCUCCUCGUUGUCGGAGGCCUCGGAACUGGCGGCCACUUCAUUUGUGGGCUCCGGCUCUUUCC --.......-----.......((((.(((..----(((....(((.---(((((........)))))....)))...)))(.((((.......)))).)...))).)))).. ( -28.80) >consensus __AUGACUU_____UAUAUCA___CUGUCAAUGGGUCUUACCUCCUCCUCCUCCUCGUUUUCGGAAGCCUCGGAACUGGCCACCACUUCAUUUGUGGGCUCCGAAUCCUUUC ...........................................................((((((((((........)))..((((.......)))).)))))))....... (-16.32 = -16.65 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:10:00 2006