| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,274,041 – 13,274,146 |
| Length | 105 |
| Max. P | 0.519898 |

| Location | 13,274,041 – 13,274,146 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -24.52 |
| Consensus MFE | -13.22 |
| Energy contribution | -12.97 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
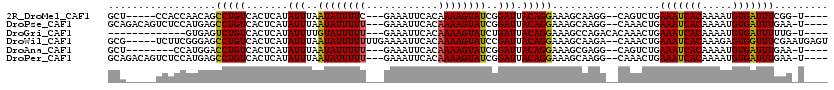
>2R_DroMel_CAF1 13274041 105 - 20766785 GCU-----CCACCAACAGCCUGUCACUCAUAUUUAAUAUUUUC---GAAAUUCACAAAAGUAUCGGAUUACAGGAAAGCAAGG--CAGUCUGAAAUCACAAAAUGUGAUUUCGG-U---- ...-----..(((....((((.((.((...(((..(((((((.---(.......).)))))))..)))...)))).....)))--).....((((((((.....))))))))))-)---- ( -24.60) >DroPse_CAF1 1268 110 - 1 GCAGACAGUCUCCAUGAGCCUGUCACUCAUAUUUAAUAUUUUU---GAAAUUCACAAAAGUAUCGGAUUACAGGAAAGCAAGG--CAAACUGAAAUCACAAAAUGUGAUUUGAA-U---- ((.(((((.(((...))).))))).((...(((..((((((((---(.......)))))))))..)))...))....))....--.....(.(((((((.....))))))).).-.---- ( -26.60) >DroGri_CAF1 1506 99 - 1 -------------GUGAGUCUGUCACUCAUAUUUUGUAUUUUU---GAAAUUCACAAAAGUAUCUGAUUACAGGAAAGCCAGACACAAACUGAAAUCACAAAAUGUGAUUUUUG-U---- -------------.((.(((((.(..((..(((..((((((((---(.......)))))))))..))).....))..).))))).)).((.((((((((.....)))))))).)-)---- ( -22.00) >DroWil_CAF1 5184 113 - 1 GCG-----UCUUCGGGAGCCUGUCACUCAUAUUUAAUAUUUUUUUGAAAAUUCACAAAAGUAUCCGAUUACAGGAAAGCAAGA--CAAACUGAAAUCACAAAGAGUGGUUUCGAAUGAGU ((.-----.(....)..)).....((((((......(((((((.(((....))).)))))))(((.......)))........--.....(((((((((.....))))))))).)))))) ( -25.50) >DroAna_CAF1 1220 102 - 1 GCU--------CCAUGGACCUGUCACUCAUAUUUAAUAUUUUU---GAAAUUCACAAAAGUAUCGGAUUACAGGAAAGCGAGG--CAGUCUGAAAUCACAAAAUGUGAUUUGAA-U---- ((.--------.(.....(((((.......(((..((((((((---(.......)))))))))..))).))))).....)..)--)....(.(((((((.....))))))).).-.---- ( -21.80) >DroPer_CAF1 1317 110 - 1 GCAGACAGUCUCCAUGAGCCUGUCACUCAUAUUUAAUAUUUUU---GAAAUUCACAAAAGUAUCGGAUUACAGGAAAGCAAGG--CAAACUGAAAUCACAAAAUGUGAUUUGAA-U---- ((.(((((.(((...))).))))).((...(((..((((((((---(.......)))))))))..)))...))....))....--.....(.(((((((.....))))))).).-.---- ( -26.60) >consensus GCA_____UCUCCAUGAGCCUGUCACUCAUAUUUAAUAUUUUU___GAAAUUCACAAAAGUAUCGGAUUACAGGAAAGCAAGG__CAAACUGAAAUCACAAAAUGUGAUUUGAA_U____ ..................(((((.......(((..((((((((............))))))))..))).)))))..................(((((((.....)))))))......... (-13.22 = -12.97 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:47 2006