| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,262,877 – 13,262,968 |
| Length | 91 |
| Max. P | 0.859110 |

| Location | 13,262,877 – 13,262,968 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 76.47 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.28 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13262877 91 - 20766785 CUAGCCAGUGGAGCUGCCAGCUAUCAUUUGAAAUUCAAGUAAAGUUGGCAGCCCCCU----GGCGUCUGAAAAAUAGCGCG-AAG--UGAAGUAA----ACU ...(((((.((.((((((((((...(((((.....)))))..)))))))))).))))----))).................-...--........----... ( -34.70) >DroSec_CAF1 2044 91 - 1 CUAAACAGUGUAGCUGCCAGCUGUCAUUUAAAAUUCAAGUAAAGUUGGCAGCCCCUU----GGCGUCUGAAAAAUAGCGCG-AAA--GGAAGAAA----ACU ............((((((((((...((((.......))))..)))))))))).((((----.(((.(((.....)))))).-.))--))......----... ( -26.10) >DroSim_CAF1 1839 91 - 1 CUAAACAGUGUAGCUGCCAGCUGUCAUUUAAAAUUCAAGUAAAGUUGGCAGCCCCUU----GGCGUCUGAAAAAUAGCGCG-AAA--GGAAGAAA----ACU ............((((((((((...((((.......))))..)))))))))).((((----.(((.(((.....)))))).-.))--))......----... ( -26.10) >DroEre_CAF1 2044 93 - 1 CUAAGCAGUGUUGCUGCCUGCUGUCAUUUUAAAUUCAAGUAAAGUUGGCAGCCCCCU----AGCGUCUGAAAAAUUGCGCGGAAA--GGAAAAAAA---UCU ....((((.....))))..((((((((((((........))))).)))))))..(((----.((((..........))))....)--)).......---... ( -21.40) >DroYak_CAF1 2051 91 - 1 CUGAGCAGUGUAGUUGUCUUCUGUCAUUUUAAAUUCAACUGAAGUUGGCAGCCCCCU----GGCGUCUGAAAAUUAGCGCG-AAA--GGAAGAAA----ACU ...........((((.(((((((((((((((........))))).))))))...(((----.(((.((((...))))))).-..)--)))))).)----))) ( -19.20) >DroAna_CAF1 2091 93 - 1 ----GCACUGCAA----CAAUUGUCAUUCAUUAUUUUUCAAAAGUUGGCAGUGCUCUCGGAAGCGUCCAAAAAAUAGCGCG-AAGGAGGACAUAAGUUUGCA ----(((((((((----(..(((...............)))..))).)))))))((((....((((..........)))).-...))))............. ( -20.66) >consensus CUAAGCAGUGUAGCUGCCAGCUGUCAUUUAAAAUUCAAGUAAAGUUGGCAGCCCCCU____GGCGUCUGAAAAAUAGCGCG_AAA__GGAAGAAA____ACU ............((((((((((....................))))))))))..........((((..........))))...................... (-14.86 = -15.28 + 0.42)

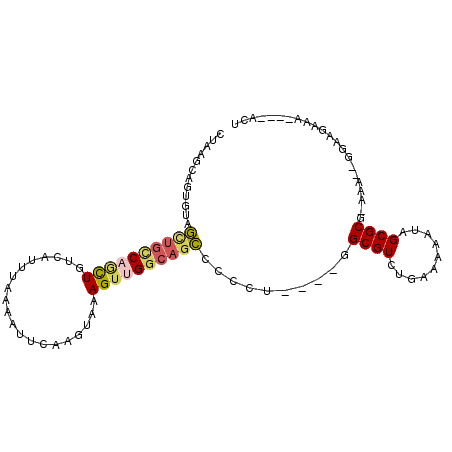

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:43 2006