
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,257,021 – 13,257,138 |
| Length | 117 |
| Max. P | 0.768991 |

| Location | 13,257,021 – 13,257,138 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -17.16 |
| Energy contribution | -18.35 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13257021 117 - 20766785 ---AAUAUUUACCGCAUCUUUUCCGAAGAUUUCGUGCACUGCGUAGACAUAAAAGUCAAUGAGCUCCAGGAUGCAAGCCACAAUCUUGGGAGAGAUGCAGUGCAAUAACUUGCACAUCUG ---..........(((((((((((.((((((..(((...(((((.(((......)))..((.....))..)))))...)))))))))))))))))))).((((((....))))))..... ( -37.90) >DroVir_CAF1 2199 120 - 1 AAUCCUUCUUACCGCAUCCCUGCCGAAUAUCUCAUGCACUGCAUACGCAUAAAAAUCAAGCAACUCCAGCAUGCACGAUACUAUCUUUGGCGAGAUGCAGUGCAGCAGCUUGCACAUUUG .............(((...((((.((.....)).((((((((((.(((...........(((.........))).(((........))))))..))))))))))))))..)))....... ( -31.50) >DroWil_CAF1 1938 120 - 1 UGAUUCACUUACCGCAUCUUUCCCAAAGAUCUCAUGCACUGCAUAGGCAUAAAAAUCAAUCAACUCCAGCAUGCAAGAGACAAUUUUCGGUGAGAUGCAAUGUAACAGUUUGCACAUCUG .............(((((((.((.((((.((((.((((.(((..((.................))...))))))).))))...)))).)).)))))))..(((((....)))))...... ( -25.83) >DroMoj_CAF1 2355 120 - 1 AUCUCUACUUACAGCAUCCUUGCCAAAGAUCUCAUGCACUGCAUAGGCAUAAAAGUCGAGCAGCUCCAGCAUGCUCGCCACUAUCUUGGGCGAGAUGCAGUGCAGCAGCUUGCACAUCUG .............(((((.(((((.(((((...((((.((....))))))....(.((((((((....)).)))))).)...))))).)))))))))).((((((....))))))..... ( -41.10) >DroAna_CAF1 2228 115 - 1 -----CACUUACUGCAUCUUUGCCAAAGAUUUCAUGCACAGCAUAAACAUAAAAAUCAAUCAACUCCAGCAUGCAGGAAACAAUCUUGGGAGAAAUACAGUGCAACAGUUUGCACAUUUG -----.......((.((.(((.((((.((((.(((((...............................)))))..(....))))))))).))).)).))((((((....))))))..... ( -19.88) >DroPer_CAF1 2348 117 - 1 ---CUUACUUACCGCGUCCUUGCCAAAGAUCUCGUGAACAGCGAACGCGUAAAAAUCAAUAAGCUCCAGCAUGCAGGACACAAUCUUAGGAGAGAUGCAGUGCAGAAGUUUGCACAUUUG ---..........(((((.(..((.(((((...(((..(.((....))..............((........)).)..))).))))).))..)))))).((((((....))))))..... ( -27.00) >consensus ___CCUACUUACCGCAUCCUUGCCAAAGAUCUCAUGCACUGCAUAGGCAUAAAAAUCAAUCAACUCCAGCAUGCAAGACACAAUCUUGGGAGAGAUGCAGUGCAACAGCUUGCACAUCUG .............(((((....((.(((((.((.((((.(((..........................))))))).))....))))).))...))))).((((((....))))))..... (-17.16 = -18.35 + 1.20)

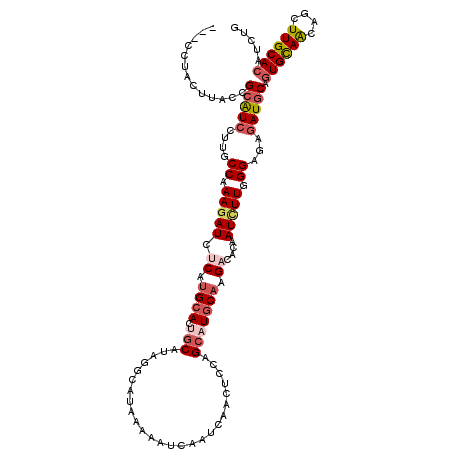
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:41 2006