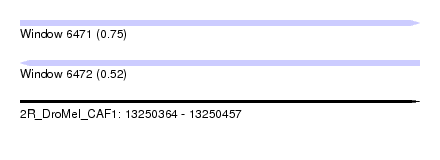
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,250,364 – 13,250,457 |
| Length | 93 |
| Max. P | 0.753654 |

| Location | 13,250,364 – 13,250,457 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -20.20 |
| Energy contribution | -21.32 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13250364 93 + 20766785 GCUGUCCACGCUGAAACUGUUGCUGCCGUUCUCGUUGAUCAGCGAGAGCGUCUGCAGGGGCAACACUUCGCCGGUGG---GUGGCUGUUCCGCGCU ((..(((((((.(((..((((.((((((((((((((....))))))))))...)))).))))....)))))..))))---)..))........... ( -38.90) >DroVir_CAF1 2139 90 + 1 GCUAUCCAUGCUGAAGCUGUUGCUGCCAUUCUCAUUGAUCAGCGAGAGCGUCUGCAGAGGCAGCAUGUC---GGUGG---GUUGCGGCUCGACAAC (((..(..(((.((.(((.((((((....((.....)).)))))).))).)).)))..)..))).((((---(((.(---....).)).))))).. ( -33.70) >DroPse_CAF1 3748 96 + 1 GCUGUCAAUACUGAAGCUGUUGCUGCCGUUCUCAUUAAUCAGCGAGAGCGUCUGCAUUGGCAGCAUAUCGGAGGGUGGUGGUGGCUGCUCCGCAUU ((..(((....)))(((.((..(..(((..(((........((....))(.((((....)))))......)))..)))..)..)).)))..))... ( -36.00) >DroGri_CAF1 1670 90 + 1 GCUUUCGAUGCUAAAGCUAUUGCUGCCAUUUUCAUUGAUCAGCGAAAGCGUUUGCAUUGGCAGCAUGUC---GGUGG---GUUGCGUAUCCGCAAC (((.(((((((....(((.((((((.((.......))..)))))).)))....))))))).))).....---.((((---((.....))))))... ( -32.00) >DroMoj_CAF1 1890 90 + 1 ACUAUCGAUGCUAAAGCUGUUGCUGCCAUUCUCGUUGAUCAGCGAGAGCGUCUGCAUGGGCAGCAGAUC---GGUGG---AUUGCGGCUCUGCAAC .((((((((((....))..((((((((..(((((((....)))))))((....))...)))))))))))---)))))---.(((((....))))). ( -37.70) >DroPer_CAF1 3284 96 + 1 GCUGUCAAUACUGAAGCUGUUGCUGCCGUUCUCAUUAAUCAGCGAGAGCGUCUGCAUUGGCAGCAUAUCGGAGGGUGGUGGUGGCUGCUCCGCAUU ((..(((....)))(((.((..(..(((..(((........((....))(.((((....)))))......)))..)))..)..)).)))..))... ( -36.00) >consensus GCUGUCAAUGCUGAAGCUGUUGCUGCCAUUCUCAUUGAUCAGCGAGAGCGUCUGCAUUGGCAGCAUAUC___GGUGG___GUGGCGGCUCCGCAAC (((.(((((((.((.(((.((((((..(((......))))))))).))).)).))))))).)))................((((.....))))... (-20.20 = -21.32 + 1.11)

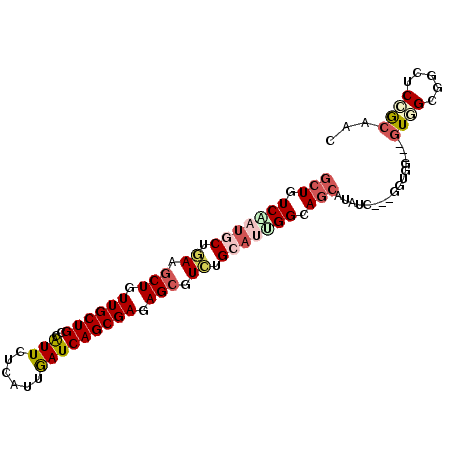
| Location | 13,250,364 – 13,250,457 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -21.59 |
| Energy contribution | -20.32 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.39 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13250364 93 - 20766785 AGCGCGGAACAGCCAC---CCACCGGCGAAGUGUUGCCCCUGCAGACGCUCUCGCUGAUCAACGAGAACGGCAGCAACAGUUUCAGCGUGGACAGC .(((((((((.(((..---.....)))....(((((((...((....))(((((........)))))..)))))))...))))).))))....... ( -31.50) >DroVir_CAF1 2139 90 - 1 GUUGUCGAGCCGCAAC---CCACC---GACAUGCUGCCUCUGCAGACGCUCUCGCUGAUCAAUGAGAAUGGCAGCAACAGCUUCAGCAUGGAUAGC (((((......)))))---(((..---....(((((((...((....))(((((........)))))..)))))))...((....)).)))..... ( -24.40) >DroPse_CAF1 3748 96 - 1 AAUGCGGAGCAGCCACCACCACCCUCCGAUAUGCUGCCAAUGCAGACGCUCUCGCUGAUUAAUGAGAACGGCAGCAACAGCUUCAGUAUUGACAGC ....(((((..............)))))....((((.((((((...((.(((((........))))).))(.(((....))).).)))))).)))) ( -25.34) >DroGri_CAF1 1670 90 - 1 GUUGCGGAUACGCAAC---CCACC---GACAUGCUGCCAAUGCAAACGCUUUCGCUGAUCAAUGAAAAUGGCAGCAAUAGCUUUAGCAUCGAAAGC ((((((....))))))---....(---((..((((((((..((....))(((((........))))).))))))))...((....)).)))..... ( -28.00) >DroMoj_CAF1 1890 90 - 1 GUUGCAGAGCCGCAAU---CCACC---GAUCUGCUGCCCAUGCAGACGCUCUCGCUGAUCAACGAGAAUGGCAGCAACAGCUUUAGCAUCGAUAGU (((((......)))))---....(---(((.(((((((...((....))(((((........)))))..)))))))...((....))))))..... ( -26.20) >DroPer_CAF1 3284 96 - 1 AAUGCGGAGCAGCCACCACCACCCUCCGAUAUGCUGCCAAUGCAGACGCUCUCGCUGAUUAAUGAGAACGGCAGCAACAGCUUCAGUAUUGACAGC ....(((((..............)))))....((((.((((((...((.(((((........))))).))(.(((....))).).)))))).)))) ( -25.34) >consensus AUUGCGGAGCAGCAAC___CCACC___GAUAUGCUGCCAAUGCAGACGCUCUCGCUGAUCAAUGAGAACGGCAGCAACAGCUUCAGCAUCGACAGC ..((((((((............(....)...(((((((...((....))(((((........)))))..)))))))...))))).)))........ (-21.59 = -20.32 + -1.27)

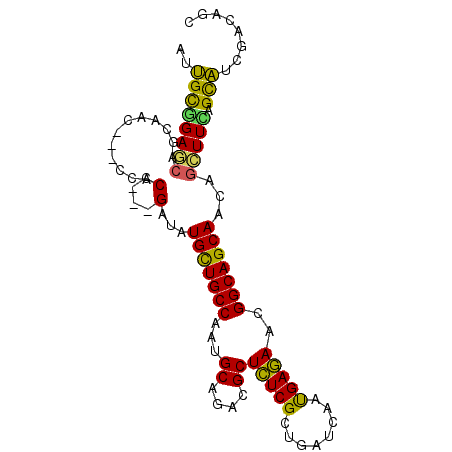
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:37 2006