| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,396,689 – 2,396,821 |
| Length | 132 |
| Max. P | 0.915718 |

| Location | 2,396,689 – 2,396,789 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -21.07 |
| Energy contribution | -19.97 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
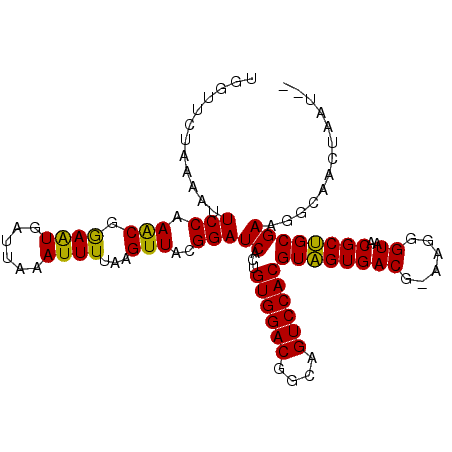
>2R_DroMel_CAF1 2396689 100 + 20766785 -----UUAAAAUUCCAAGCGGAGUGAUUACAUUUUAAGUUAAGGAUCAAUGUGGACAGCAGUCCACGUUGUGACGGAAGGGUAACGCAGCGAAGGUAGGUUGUCC -----......((((.(((.(((((....)))))...)))..(...((((((((((....))))))))))...)))))((..(((.(.((....)).))))..)) ( -30.60) >DroSec_CAF1 52428 102 + 1 UGGUUCUAAAAUUCCAAACGGAAUGAUUAAAUUUUAAGUUACGGAUCACUGUGGACGGUAGUCCACGUAGUGACG-AAGGGUAACGCUGCGAAGGCAACUAAU-- (((((.....(((((....))))).............(((((...(((((((((((....))))))..)))))(.-...))))))...((....)))))))..-- ( -29.60) >DroSim_CAF1 9808 102 + 1 UGGUUCUAAAAUUCCAAACGAAAUGAUUAAAUUUUAAGUUACGGAUCACUGUGGACGGCAGUCCACGUAGUGACG-AAGGGUAACGCUGCGAAGGCAACUAAU-- (((..........)))...(((((......)))))..(((((...(((((((((((....))))))..)))))(.-...))))))..(((....)))......-- ( -26.30) >consensus UGGUUCUAAAAUUCCAAACGGAAUGAUUAAAUUUUAAGUUACGGAUCACUGUGGACGGCAGUCCACGUAGUGACG_AAGGGUAACGCUGCGAAGGCAACUAAU__ ............(((.(((.((((......))))...)))..)))((...((((((....))))))((((((((......))..))))))))............. (-21.07 = -19.97 + -1.10)

| Location | 2,396,689 – 2,396,789 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 84.62 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -17.44 |
| Energy contribution | -19.33 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792019 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2396689 100 - 20766785 GGACAACCUACCUUCGCUGCGUUACCCUUCCGUCACAACGUGGACUGCUGUCCACAUUGAUCCUUAACUUAAAAUGUAAUCACUCCGCUUGGAAUUUUAA----- ((((((..............((.((......)).))...((((((....)))))).))).)))....................(((....))).......----- ( -16.40) >DroSec_CAF1 52428 102 - 1 --AUUAGUUGCCUUCGCAGCGUUACCCUU-CGUCACUACGUGGACUACCGUCCACAGUGAUCCGUAACUUAAAAUUUAAUCAUUCCGUUUGGAAUUUUAGAACCA --....(((((....)))))(((((....-.((((((..((((((....))))))))))))..)))))......(((((..(((((....))))).))))).... ( -29.10) >DroSim_CAF1 9808 102 - 1 --AUUAGUUGCCUUCGCAGCGUUACCCUU-CGUCACUACGUGGACUGCCGUCCACAGUGAUCCGUAACUUAAAAUUUAAUCAUUUCGUUUGGAAUUUUAGAACCA --....(((((....)))))(((((....-.((((((..((((((....))))))))))))..)))))(((((((((...(.....)....)))))))))..... ( -27.00) >consensus __AUUAGUUGCCUUCGCAGCGUUACCCUU_CGUCACUACGUGGACUGCCGUCCACAGUGAUCCGUAACUUAAAAUUUAAUCAUUCCGUUUGGAAUUUUAGAACCA ......(((((....)))))(((((......((((((..((((((....))))))))))))..))))).............(((((....))))).......... (-17.44 = -19.33 + 1.89)

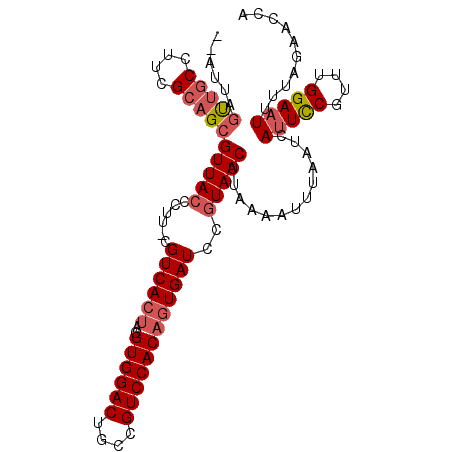
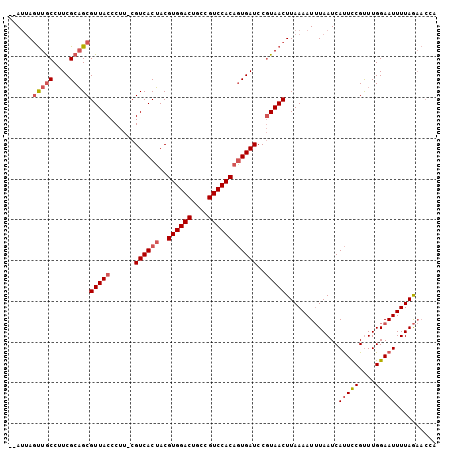
| Location | 2,396,722 – 2,396,821 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -19.21 |
| Energy contribution | -19.10 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543734 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2396722 99 + 20766785 UUAAGGAUCAAUGUGGACAGCAGUCCACGUUGUGACGGAAGGGUAACGCAGCGAAGGUAGGUUGUCCAGUAGGUUGGUAAUAGGGAGUAGAAG-----UGGUAU-- ......((((((.((((((((...((.(((((((.(....).....)))))))..))...))))))))....))))))...............-----......-- ( -27.40) >DroSec_CAF1 52466 103 + 1 UUACGGAUCACUGUGGACGGUAGUCCACGUAGUGACG-AAGGGUAACGCUGCGAAGGCAACUAAU--AAUAGGUAGGUAAUAGGUAGUAGUAGGUGACUGGUAGGC ((((..(((...((((((....))))))((((((.(.-...)....))))))..((....))...--....)))..)))).......((.(((....))).))... ( -28.90) >DroSim_CAF1 9846 103 + 1 UUACGGAUCACUGUGGACGGCAGUCCACGUAGUGACG-AAGGGUAACGCUGCGAAGGCAACUAAU--AAUAGGUAGGUAAUAGGUAGUAGUAGGUGACUGGUGGGC ((((..(((...((((((....))))))((((((.(.-...)....))))))..((....))...--....)))..)))).......((.(((....))).))... ( -28.60) >consensus UUACGGAUCACUGUGGACGGCAGUCCACGUAGUGACG_AAGGGUAACGCUGCGAAGGCAACUAAU__AAUAGGUAGGUAAUAGGUAGUAGUAGGUGACUGGUAGGC ((((..(((...((((((....))))))((((((((......))..))))))...................)))..)))).......................... (-19.21 = -19.10 + -0.11)

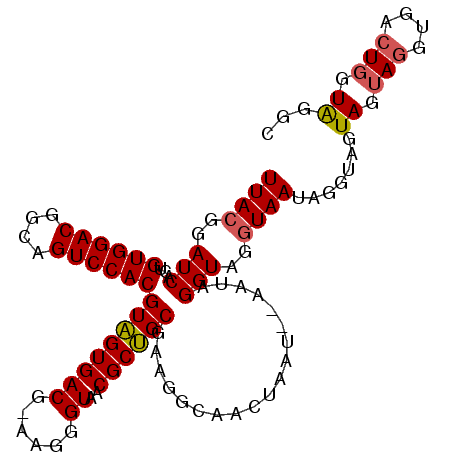
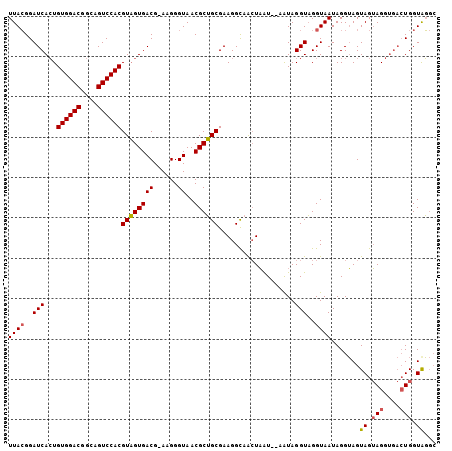
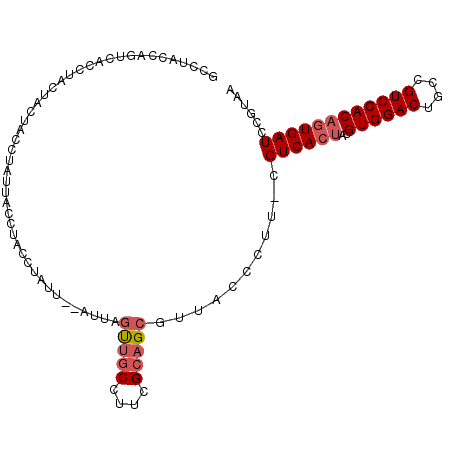
| Location | 2,396,722 – 2,396,821 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -19.53 |
| Consensus MFE | -14.25 |
| Energy contribution | -15.70 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2396722 99 - 20766785 --AUACCA-----CUUCUACUCCCUAUUACCAACCUACUGGACAACCUACCUUCGCUGCGUUACCCUUCCGUCACAACGUGGACUGCUGUCCACAUUGAUCCUUAA --......-----..........................((((((..............((.((......)).))...((((((....)))))).))).))).... ( -14.80) >DroSec_CAF1 52466 103 - 1 GCCUACCAGUCACCUACUACUACCUAUUACCUACCUAUU--AUUAGUUGCCUUCGCAGCGUUACCCUU-CGUCACUACGUGGACUACCGUCCACAGUGAUCCGUAA ...(((..(((((..........................--....(((((....))))).........-.........((((((....)))))).)))))..))). ( -22.40) >DroSim_CAF1 9846 103 - 1 GCCCACCAGUCACCUACUACUACCUAUUACCUACCUAUU--AUUAGUUGCCUUCGCAGCGUUACCCUU-CGUCACUACGUGGACUGCCGUCCACAGUGAUCCGUAA .................(((...................--....(((((....))))).........-.((((((..((((((....))))))))))))..))). ( -21.40) >consensus GCCUACCAGUCACCUACUACUACCUAUUACCUACCUAUU__AUUAGUUGCCUUCGCAGCGUUACCCUU_CGUCACUACGUGGACUGCCGUCCACAGUGAUCCGUAA .............................................(((((....)))))...........((((((..((((((....))))))))))))...... (-14.25 = -15.70 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:41 2006