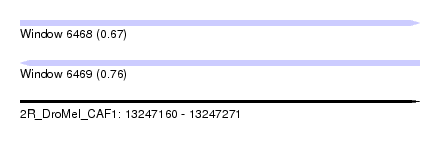
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,247,160 – 13,247,271 |
| Length | 111 |
| Max. P | 0.757634 |

| Location | 13,247,160 – 13,247,271 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -34.42 |
| Consensus MFE | -22.16 |
| Energy contribution | -21.13 |
| Covariance contribution | -1.02 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13247160 111 + 20766785 CACAUACGACGGCUCCUAAUUGGCAGUCGCUUGGUACUCCCUUUCAGACUAUAUCCGGACGCAGGCGUCCUUUGCCUGCAUAACACUGUUCGUGAUGAGCUUGGGUGCCGU ........(((((.((((...(((((((....((.....)).....)))).((((((((((((((((.....)))))))........))))).)))).))))))).))))) ( -39.80) >DroVir_CAF1 6774 111 + 1 CUAUUGUUUCAGCUUUUAUUUGGUAGUGUUAUGGCACGCCAUUGCAGAUUACAUACGAACGCAGGCAUCGUUUGCCUGCAUCACAUUGUUCGUGAUGAGCCUGGGUGCUGU .........((((.......((((.((((....))))))))...(((..(.((((((((((((((((.....)))))))........)))))).))))..)))...)))). ( -34.40) >DroGri_CAF1 6695 111 + 1 CAUAUGUUUCCGUUUUUAUUUGGUAGUGGCGUGGUAUUUCCUUUCAGAUUAUAUUAGAACGCAGGCAUCCUUUGCCUGCAUUACGUUGUUUGUCAUGAGCCUGGGCGCUGU .....((.((((..(((((.....((..(((((((.......(((.(((...))).))).(((((((.....))))))))))))))..))....)))))..)))).))... ( -27.90) >DroWil_CAF1 5110 111 + 1 CGUGGUAGACAGCUUGUAAUGGGCAGUGACUUGGUAUACCCUUUCAGAUUACAUCCGCACGCAGGCAUCCUUUGCCUGCAUCACGCUAUUUGUCAUGAGUCUGGGAGCCGU ...(((...(((((..(.((.(((.((((...((....))....................(((((((.....))))))).)))))))....)).)..)).)))...))).. ( -31.20) >DroAna_CAF1 5759 111 + 1 UUUAAGUAACAGCUCCUAAUUGGCAGUGGCUUGGUACUUCCUUUCAGACUAUAUCCGGACGCAGGCGUCCUUUGCCUGCAUUACGUUGUUCGUGAUGAGCUUGGGAGCUGU ........(((((((((((..(((((.((..((((.((.......))))))...))(((((....))))).)))))..(((((((.....)))))))...))))))))))) ( -42.10) >DroPer_CAF1 5854 106 + 1 -----UUGACAGUCCGUAAUUGGCAGUGGGUUGGUAUUUCCUUUCAGACUACAUCCGAACGCAGGCAUCGUUUGCCUGCAUCACACUGUUCGUCAUGAGCCUGGGCGCCGU -----......(((((.....((((((.((..((.....))..))..))).(((.((((((((((((.....)))))))........)))))..))).))))))))..... ( -31.10) >consensus CAUAUGUGACAGCUCCUAAUUGGCAGUGGCUUGGUACUCCCUUUCAGACUACAUCCGAACGCAGGCAUCCUUUGCCUGCAUCACACUGUUCGUCAUGAGCCUGGGAGCCGU ...........((.((((...(((.(((((..((....))....................(((((((.....)))))))............)))))..))))))).))... (-22.16 = -21.13 + -1.02)

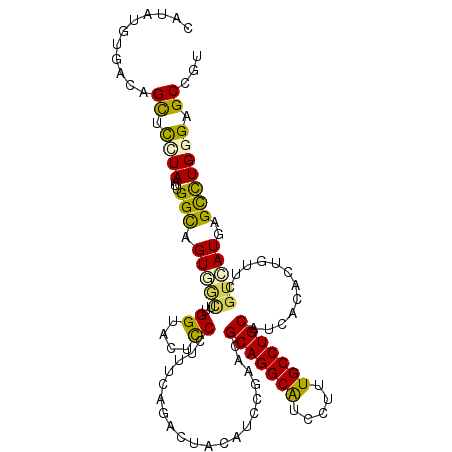
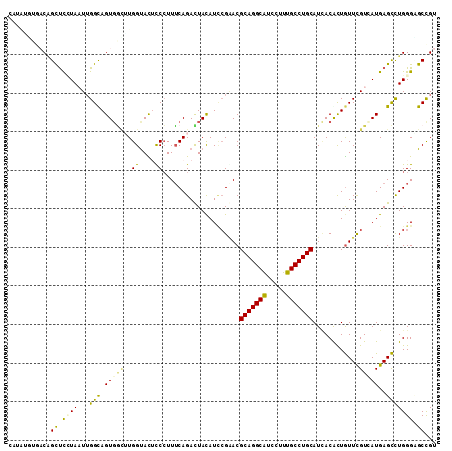
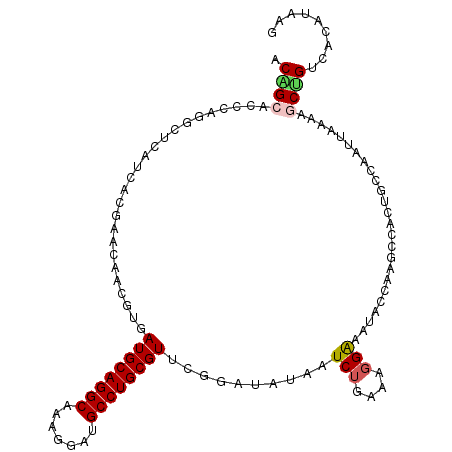
| Location | 13,247,160 – 13,247,271 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -17.08 |
| Energy contribution | -17.22 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13247160 111 - 20766785 ACGGCACCCAAGCUCAUCACGAACAGUGUUAUGCAGGCAAAGGACGCCUGCGUCCGGAUAUAGUCUGAAAGGGAGUACCAAGCGACUGCCAAUUAGGAGCCGUCGUAUGUG (((((.((...((....(((.....)))....)).((((..(((((....)))))((.(((..(((....))).))))).......)))).....)).)))))........ ( -35.10) >DroVir_CAF1 6774 111 - 1 ACAGCACCCAGGCUCAUCACGAACAAUGUGAUGCAGGCAAACGAUGCCUGCGUUCGUAUGUAAUCUGCAAUGGCGUGCCAUAACACUACCAAAUAAAAGCUGAAACAAUAG .((((...(((..((((.((((((........(((((((.....)))))))))))))))).)..)))..((((....)))).................))))......... ( -31.80) >DroGri_CAF1 6695 111 - 1 ACAGCGCCCAGGCUCAUGACAAACAACGUAAUGCAGGCAAAGGAUGCCUGCGUUCUAAUAUAAUCUGAAAGGAAAUACCACGCCACUACCAAAUAAAAACGGAAACAUAUG ..........(((..............(.((((((((((.....)))))))))))........(((....)))........)))................(....)..... ( -24.50) >DroWil_CAF1 5110 111 - 1 ACGGCUCCCAGACUCAUGACAAAUAGCGUGAUGCAGGCAAAGGAUGCCUGCGUGCGGAUGUAAUCUGAAAGGGUAUACCAAGUCACUGCCCAUUACAAGCUGUCUACCACG ((((((((((...(((((........)))))((((((((.....)))))))))).)))((((((......(((((...........))))))))))).)))))........ ( -32.10) >DroAna_CAF1 5759 111 - 1 ACAGCUCCCAAGCUCAUCACGAACAACGUAAUGCAGGCAAAGGACGCCUGCGUCCGGAUAUAGUCUGAAAGGAAGUACCAAGCCACUGCCAAUUAGGAGCUGUUACUUAAA ((((((((......(((.(((.....))).)))..((((..(((((....)))))((.(((..(((....))).))))).......)))).....))))))))........ ( -36.20) >DroPer_CAF1 5854 106 - 1 ACGGCGCCCAGGCUCAUGACGAACAGUGUGAUGCAGGCAAACGAUGCCUGCGUUCGGAUGUAGUCUGAAAGGAAAUACCAACCCACUGCCAAUUACGGACUGUCAA----- ..((.((....)))).(((((..(((((.((((((((((.....)))))))))).((.(((..(((....))).)))))....)))))((......))..))))).----- ( -33.10) >consensus ACAGCACCCAGGCUCAUCACGAACAACGUGAUGCAGGCAAAGGAUGCCUGCGUUCGGAUAUAAUCUGAAAGGAAAUACCAAGCCACUGCCAAUUAAAAGCUGUCACAUAAG .((((.........................((((((((.......))))))))..........(((....))).........................))))......... (-17.08 = -17.22 + 0.14)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:34 2006