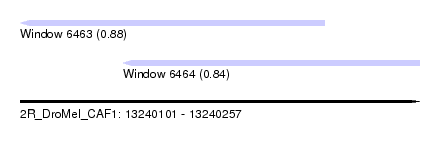
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,240,101 – 13,240,257 |
| Length | 156 |
| Max. P | 0.884354 |

| Location | 13,240,101 – 13,240,220 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.37 |
| Mean single sequence MFE | -32.39 |
| Consensus MFE | -27.76 |
| Energy contribution | -26.33 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884354 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
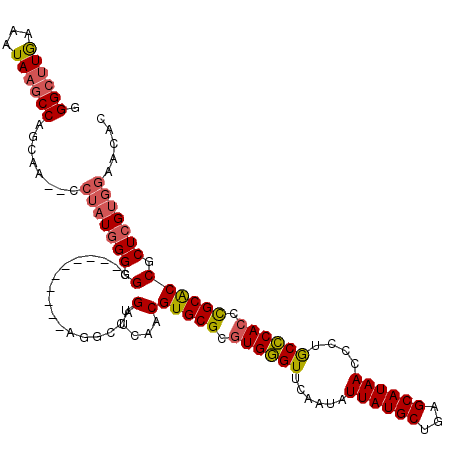
>2R_DroMel_CAF1 13240101 119 - 20766785 CCAAAAUCCAGUUGGACGUGCGUGUGCGUACAACAUUAUGCUGAGCAUAACUCUACGCAACUGCGCCGCUUG-GAUGCAAAUCCAAUACAAUAAUACAAACCAAGUGCAUAUCGAGGGGU ((......((.((((.((.((((.((((((.....((((((...))))))...))))))...)))))).(((-(((....))))))..............)))).)).........)).. ( -30.76) >DroPse_CAF1 20794 109 - 1 ----AGGCCAGUUCAACGUGCGCGUGGGUUCAAUAUUAUGCUGAGCAUAACCCUGCCCACCCGCACCGCUCGUGGAACACAUCCA------UAUCCAAAACCAAGUGCAUAUCGAGGCG- ----..(((.((((.(((((((.((((((......((((((...))))))....)))))).))))).....)).))))...((.(------(((.((........)).)))).))))).- ( -33.20) >DroPer_CAF1 20836 109 - 1 ----AGGCCAGUUCAACGUGCGCGUGGGUUCAAUAUUAUGCUGAGCAUAACCCUGCCCACCCGCACCGCUCGUGGAACACAUCCA------UAUCCAAAACCAAGUGCAUAUCGAGGCG- ----..(((.((((.(((((((.((((((......((((((...))))))....)))))).))))).....)).))))...((.(------(((.((........)).)))).))))).- ( -33.20) >consensus ____AGGCCAGUUCAACGUGCGCGUGGGUUCAAUAUUAUGCUGAGCAUAACCCUGCCCACCCGCACCGCUCGUGGAACACAUCCA______UAUCCAAAACCAAGUGCAUAUCGAGGCG_ ......(((........(((((.((((((......((((((...))))))....)))))).)))))((((..((((.................))))......))))........))).. (-27.76 = -26.33 + -1.43)


| Location | 13,240,141 – 13,240,257 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.86 |
| Mean single sequence MFE | -37.33 |
| Consensus MFE | -27.86 |
| Energy contribution | -28.87 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13240141 116 - 20766785 GGGAAUAAAGUAGACCUUCG---CCAAUCGGAGGUGAUAACCAAAAUCCAGUUGGACGUGCGUGUGCGUACAACAUUAUGCUGAGCAUAACUCUACGCAACUGCGCCGCUUG-GAUGCAA .........(((.(((((((---.....))))))).....((((..(((....)))((.((((.((((((.....((((((...))))))...))))))...)))))).)))-).))).. ( -33.50) >DroPse_CAF1 20827 110 - 1 GGGCUUGAAAUAAGCCAGCAAAUUCUAUGGGGGG----------AGGCCAGUUCAACGUGCGCGUGGGUUCAAUAUUAUGCUGAGCAUAACCCUGCCCACCCGCACCGCUCGUGGAACAC .((((((...))))))......(((((((((.((----------((.....)))...(((((.((((((......((((((...))))))....)))))).)))))).)))))))))... ( -40.40) >DroPer_CAF1 20869 110 - 1 GGGCUUGAAAUAAGCCAGCAAUAGCUAUGGGGGG----------AGGCCAGUUCAACGUGCGCGUGGGUUCAAUAUUAUGCUGAGCAUAACCCUGCCCACCCGCACCGCUCGUGGAACAC .((((((...))))))........(((((((.((----------((.....)))...(((((.((((((......((((((...))))))....)))))).)))))).)))))))..... ( -38.10) >consensus GGGCUUGAAAUAAGCCAGCAA__CCUAUGGGGGG__________AGGCCAGUUCAACGUGCGCGUGGGUUCAAUAUUAUGCUGAGCAUAACCCUGCCCACCCGCACCGCUCGUGGAACAC .((((((...))))))........(((((((.(.................(.....)(((((.((((((......((((((...))))))....)))))).)))))).)))))))..... (-27.86 = -28.87 + 1.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:28 2006