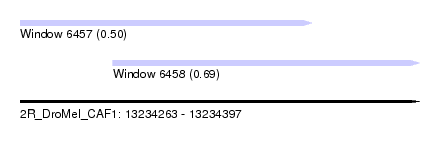
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,234,263 – 13,234,397 |
| Length | 134 |
| Max. P | 0.692521 |

| Location | 13,234,263 – 13,234,361 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 96.26 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13234263 98 + 20766785 AGAGAGCGCCAAAUCAUCCGGUGGAAAUGGACAUAUGGGUGGUUGGAUAGGGGCAGUGGGAGGGAUGAGAGUUCGUUUUCUACUGAAUGAAAGCGACA .....((.((((.((((((((((........))).))))))))))).......(((((((((((((....))))..))))))))).......)).... ( -28.80) >DroSec_CAF1 14688 98 + 1 AGAGAGCGCCAAAUCAUCCGGUGGAAAUGGACAUAUGGGUGGAUUGAAAGGGGCAGGGGGAGGGAUGAGAGUUCGUUUUCUACUGAAUGAAAGCGACA .....(((((...((((((((((........))).))))))).((....))))).((..((.((((....)))).))..))...........)).... ( -25.80) >DroSim_CAF1 15931 98 + 1 AGAGAGCGCCAAAUCAUCCGGUGGAAAUGGACAUAUGGGUGGAUGGACAGGGGCGGGGGGAGGGAUGAGAGUUCGUUUUCUACUGAAUGAAAGCGACA .....((.(((..((((((((((........))).))))))).))).(((.....((..((.((((....)))).))..)).))).......)).... ( -26.70) >consensus AGAGAGCGCCAAAUCAUCCGGUGGAAAUGGACAUAUGGGUGGAUGGAAAGGGGCAGGGGGAGGGAUGAGAGUUCGUUUUCUACUGAAUGAAAGCGACA .....((.((...((((((((((........))).))))))).......)).))................((.(((((((........))))))))). (-22.10 = -22.10 + -0.00)


| Location | 13,234,294 – 13,234,397 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.19 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -20.17 |
| Energy contribution | -21.43 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13234294 103 + 20766785 CAUAUGGGUGGUUGGAUAGGGGCAGUGGGAGGGAUGAGAGUUCGUUUUCUACUGAAUGAAAGCG-----------ACAGGCUUCGUGUGAC-UUCGUC-UGCUCUCGGCUACCCUU .....(((((((((....(((((((..((((..(((.((((((((((((........)))))))-----------)...))))..)))..)-)))..)-))))))))))))))).. ( -39.20) >DroSec_CAF1 14719 103 + 1 CAUAUGGGUGGAUUGAAAGGGGCAGGGGGAGGGAUGAGAGUUCGUUUUCUACUGAAUGAAAGCG-----------ACAGGCUUCGUGUGAC-UUCGUC-UGCACUCGUCUGCCCUU .....(((..(((.....((((((((.((((..(((.((((((((((((........)))))))-----------)...))))..)))..)-))).))-))).))))))..))).. ( -38.30) >DroSim_CAF1 15962 103 + 1 CAUAUGGGUGGAUGGACAGGGGCGGGGGGAGGGAUGAGAGUUCGUUUUCUACUGAAUGAAAGCG-----------ACAGGCUUCGUGUGAC-UUCGUC-UGUGCUCGUCUGCCCUU .....(((..((((..((.((((((((....(.((((.(((((((((((........)))))))-----------)...))))))).)..)-))))))-).))..))))..))).. ( -43.00) >DroAna_CAF1 17681 110 + 1 UAUAUGUAUGUAAUAAGAGG----GAGGGAGAAAUGUGGG--CGAUUUCUACUGAAUGAAAGCGAGUGCAGCCCGACGGGAUUUGAGUGAGAUUCGUUCCGCUUCGUUCUGAGCUU ..............(((.((----((.((((.....((((--(..((((........))))((....)).))))).((((((..((((...)))))))))))))).))))...))) ( -25.90) >consensus CAUAUGGGUGGAUGGAAAGGGGCAGGGGGAGGGAUGAGAGUUCGUUUUCUACUGAAUGAAAGCG___________ACAGGCUUCGUGUGAC_UUCGUC_UGCUCUCGUCUGCCCUU .................((((((((((((((.((((((....(((((((........)))))))............((.((...)).))...))))))...))))).))))))))) (-20.17 = -21.43 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:23 2006