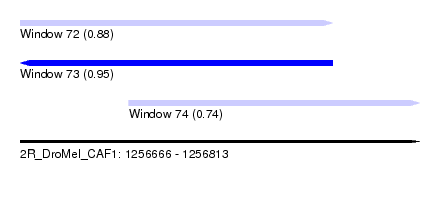
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,256,666 – 1,256,813 |
| Length | 147 |
| Max. P | 0.945589 |

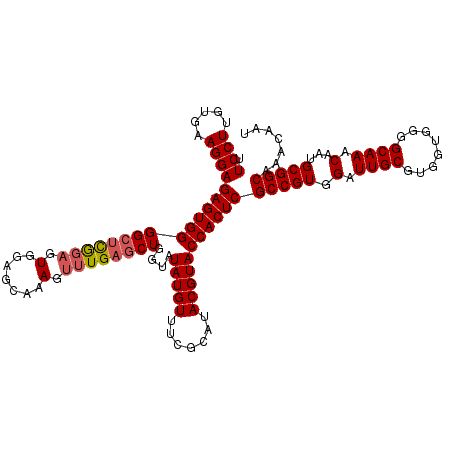
| Location | 1,256,666 – 1,256,781 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -37.83 |
| Consensus MFE | -34.34 |
| Energy contribution | -34.82 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1256666 115 + 20766785 UUCCUUGUGAAGGAGAGUGGGGCUCAGAGUGGAGCAAAGUUUGUGCUGGUAUAUGUUUCGCAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAU (((((.....)))))......((((......))))...((((((((..((...(((..(((.((((((((((.....))))..)))))))))..))).))...)).))))))... ( -37.60) >DroSec_CAF1 8356 115 + 1 UUCCUUGUGAAGGAGAGUGGGGCUCGGAGUGGAGCAAAGUUUGAGCUGGUAUAUGUUUCGCAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAU .((((.....))))((((((((((((((.(.......).)))))))).(..((((.....))))..).))))))(((((.(.((((........)))).)...)))))....... ( -38.10) >DroSim_CAF1 14167 115 + 1 UUCCUUGUGAAGGAGAGUGGGGCUCGGAGUGGAGCAAAGUUUGAGCUGGUAUAUGUUUCGCAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAU .((((.....))))((((((((((((((.(.......).)))))))).(..((((.....))))..).))))))(((((.(.((((........)))).)...)))))....... ( -38.10) >DroEre_CAF1 12495 115 + 1 UUCCUUAUGAAGGAGAGUGGGGCUUGGAGUGGACCAAAGUUUGAGCUGGUAUAUGUUUCGUAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAU .((((.....))))((((((((((..((.(.......).))..)))).(((((((...)))))))...))))))(((((.(.((((........)))).)...)))))....... ( -39.80) >DroYak_CAF1 11848 115 + 1 UUCCUUAUAAAGGAGAGUGGGGCUCGCAGUGGACCAAAGAUUGAACUGGUAUAUGUUUCGUAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAU .((((.....))))((((((..(...((((..............))))(((((((...))))))))..))))))(((((.(.((((........)))).)...)))))....... ( -35.54) >consensus UUCCUUGUGAAGGAGAGUGGGGCUCGGAGUGGAGCAAAGUUUGAGCUGGUAUAUGUUUCGCAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAU .((((.....))))((((((((((((((.(.......).))))))))....(((((.......)))))))))))(((((.(.((((........)))).)...)))))....... (-34.34 = -34.82 + 0.48)

| Location | 1,256,666 – 1,256,781 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -24.75 |
| Energy contribution | -24.75 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945589 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1256666 115 - 20766785 AUUGUUUGCCGCAUUGUUUGCCCCACCACGCAAUCCACGGCGAGUGGUACGUAUGCGAAACAUAUACCAGCACAAACUUUGCUCCACUCUGAGCCCCACUCUCCUUCACAAGGAA .......((((.(((((.((......)).)))))...))))((((((...(((((.....)))))...............((((......)))).))))))((((.....)))). ( -28.50) >DroSec_CAF1 8356 115 - 1 AUUGUUUGCCGCAUUGUUUGCCCCACCACGCAAUCCACGGCGAGUGGUACGUAUGCGAAACAUAUACCAGCUCAAACUUUGCUCCACUCCGAGCCCCACUCUCCUUCACAAGGAA .......((((.(((((.((......)).)))))...))))((((((...(((((.....)))))...............((((......)))).))))))((((.....)))). ( -28.60) >DroSim_CAF1 14167 115 - 1 AUUGUUUGCCGCAUUGUUUGCCCCACCACGCAAUCCACGGCGAGUGGUACGUAUGCGAAACAUAUACCAGCUCAAACUUUGCUCCACUCCGAGCCCCACUCUCCUUCACAAGGAA .......((((.(((((.((......)).)))))...))))((((((...(((((.....)))))...............((((......)))).))))))((((.....)))). ( -28.60) >DroEre_CAF1 12495 115 - 1 AUUGUUUGCCGCAUUGUUUGCCCCACCACGCAAUCCACGGCGAGUGGUACGUAUACGAAACAUAUACCAGCUCAAACUUUGGUCCACUCCAAGCCCCACUCUCCUUCAUAAGGAA .......((((.(((((.((......)).)))))...))))((((((..((((((.......)))))...........((((......)))))..))))))((((.....)))). ( -27.60) >DroYak_CAF1 11848 115 - 1 AUUGUUUGCCGCAUUGUUUGCCCCACCACGCAAUCCACGGCGAGUGGUACGUAUACGAAACAUAUACCAGUUCAAUCUUUGGUCCACUGCGAGCCCCACUCUCCUUUAUAAGGAA .......((((.(((((.((......)).)))))...))))((((((..((....))........(((((........)))))............))))))((((.....)))). ( -26.30) >consensus AUUGUUUGCCGCAUUGUUUGCCCCACCACGCAAUCCACGGCGAGUGGUACGUAUGCGAAACAUAUACCAGCUCAAACUUUGCUCCACUCCGAGCCCCACUCUCCUUCACAAGGAA .......((((.(((((.((......)).)))))...))))((((((...(((((.......)))))..((((.................)))).))))))((((.....)))). (-24.75 = -24.75 + -0.00)

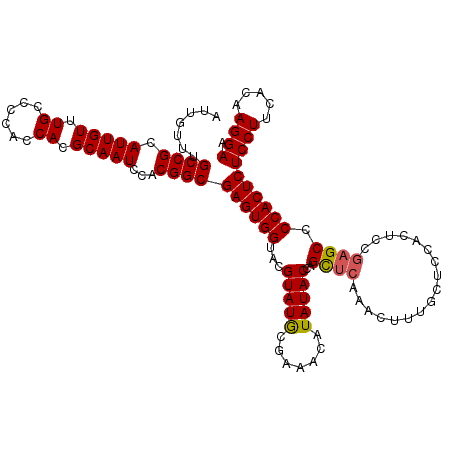
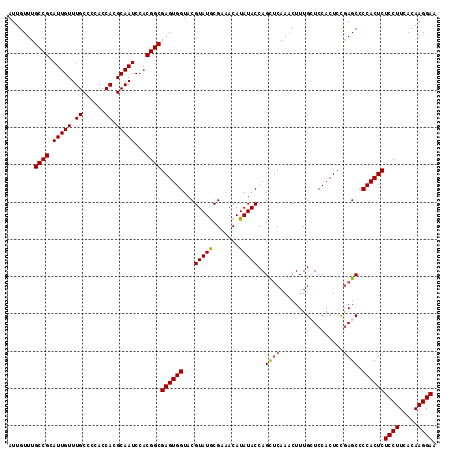
| Location | 1,256,706 – 1,256,813 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 97.76 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -34.86 |
| Energy contribution | -35.02 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1256706 107 + 20766785 UUGUGCUGGUAUAUGUUUCGCAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAUGGUUGCCGCUGAUGCCACUGCAGAUGACUUUG ((((..((((((.(((..(((.((((((((((.....))))..)))))))))..)))......(((((((.......)))))))..))))))..))))......... ( -40.80) >DroSec_CAF1 8396 107 + 1 UUGAGCUGGUAUAUGUUUCGCAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAUGGUCGCCGCUGAUGCCACUGCAGAUGACUUUG (((((.((((((.(((..(((.((((((((((.....))))..)))))))))..)))......(((((...........)))))..)))))))).)))......... ( -36.80) >DroSim_CAF1 14207 107 + 1 UUGAGCUGGUAUAUGUUUCGCAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAUGGUCGCCGCUGAUGCCACUGCAGAUGACUUUG (((((.((((((.(((..(((.((((((((((.....))))..)))))))))..)))......(((((...........)))))..)))))))).)))......... ( -36.80) >DroEre_CAF1 12535 107 + 1 UUGAGCUGGUAUAUGUUUCGUAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAUGGUUGCCGCUGAUGCCACUGCAGAUGACUCUG ...((.((((((.(((..(...((((((((((.....))))..))))))..)..)))......(((((((.......)))))))..)))))))).((((....)))) ( -35.70) >DroYak_CAF1 11888 107 + 1 UUGAACUGGUAUAUGUUUCGUAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAUGGUUGCCGCUGAUGCCACUGCAGAUGACUUUG (((...((((((.(((..(...((((((((((.....))))..))))))..)..)))......(((((((.......)))))))..))))))...)))......... ( -34.50) >consensus UUGAGCUGGUAUAUGUUUCGCAUACGUACCACUCGCCGUGGAUUGCGUGGUGGGGCAAACAAUGCGGCAAACAAUGGUUGCCGCUGAUGCCACUGCAGAUGACUUUG (((...((((((.(((..(((.((((((((((.....))))..)))))))))..)))......(((((((.......)))))))..))))))...)))......... (-34.86 = -35.02 + 0.16)

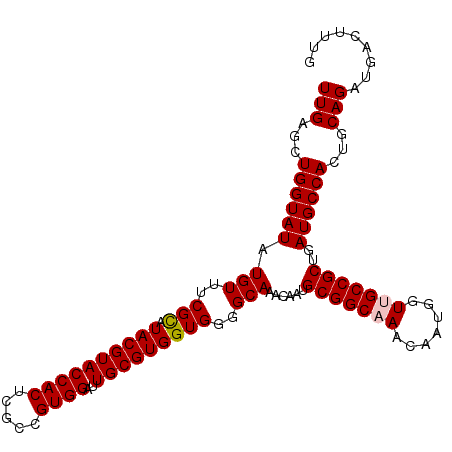
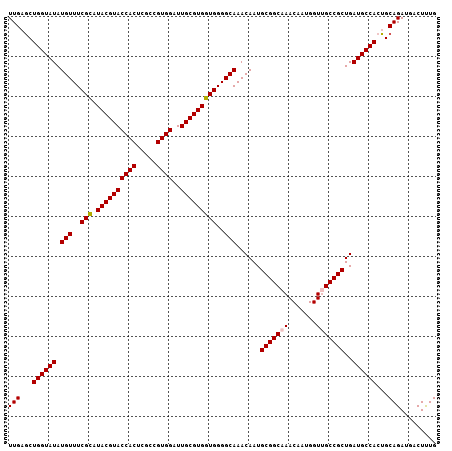
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:26 2006