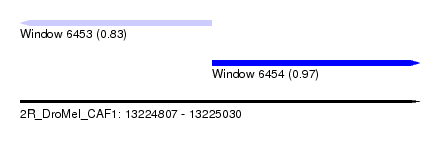
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,224,807 – 13,225,030 |
| Length | 223 |
| Max. P | 0.968218 |

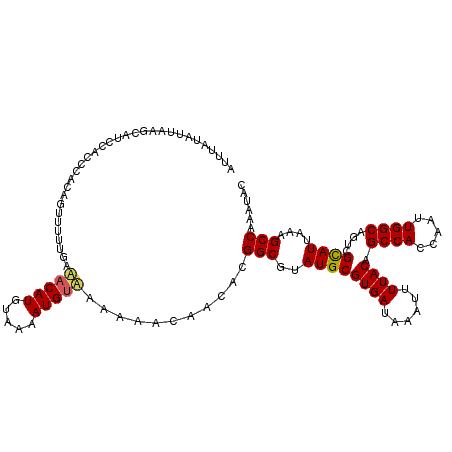
| Location | 13,224,807 – 13,224,914 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -26.60 |
| Consensus MFE | -20.95 |
| Energy contribution | -22.40 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13224807 107 - 20766785 GUUUGCAGCACUAUAACGGCAGUCUCAUAGGAUUUAUUUGAAAUGGAGUGUGUAUUUCAAAUACUUAUU-------------AUUACUUGCUUUCUGAGUGUAUAAUAUUGCAAAUAUAU ((((((((((((.....((((((...(((((...(((((((((((.......)))))))))))))))).-------------...)).)))).....))))).......))))))).... ( -23.91) >DroSec_CAF1 5421 120 - 1 GUUUGCAGCAUUAUAACGGCAGUCUGUUAAGAUUUAUUUGAAAUGGAGUGUGUAUUUCAAAUAGAUUUCGAACUCAAAGGAACUUAUGUAUUUUCUGAGUGCAUAAUAUUGUAAAUAUAU ((((((((.((((((((((....))))))((((((((((((((((.......)))))))))))))))).(.(((((.((((((....)).)))).))))).).)))).)))))))).... ( -27.90) >DroSim_CAF1 6359 120 - 1 GUUUGCAGCAUUGUAACGGCAGUCUGUUAAGAUUUAUUUGAAAUGGAGUGUGUAUUUCAAAUAGAUUUCGAACUCAAAAGAACUUAUGUAUUUUCUGAGUGCAUAAUAUUGUAAAUAUAU ((((((((.((((((((((....))))).((((((((((((((((.......)))))))))))))))).(.(((((.((((((....)).)))).))))).)))))).)))))))).... ( -28.00) >consensus GUUUGCAGCAUUAUAACGGCAGUCUGUUAAGAUUUAUUUGAAAUGGAGUGUGUAUUUCAAAUAGAUUUCGAACUCAAA_GAACUUAUGUAUUUUCUGAGUGCAUAAUAUUGUAAAUAUAU ((((((((.....((((((....))))))((((((((((((((((.......))))))))))))))))...............((((((((((...))))))))))..)))))))).... (-20.95 = -22.40 + 1.45)

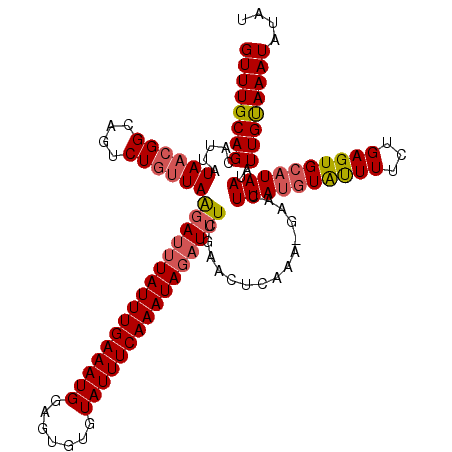
| Location | 13,224,914 – 13,225,030 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -23.10 |
| Consensus MFE | -18.04 |
| Energy contribution | -18.27 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13224914 116 + 20766785 AUUUAUAUUAAGCAUCCACCCACAGUUUUUGAAUCAUGUAAAAUGUUAAAAGCAACACGGCGUAUGCGUGAUACACUUUACAGCCACCAAUUGGCAGUCGUAUUAAUGCCAAAUAC .....((((..((((.........((((((((..(((.....)))))))))))...(((((......((((......)))).((((.....)))).)))))....))))..)))). ( -23.20) >DroSec_CAF1 5541 94 + 1 AUUUAUAUUA----------------------UACAUGUAAAAUGUAAAAAACAACACGGCGUAUGCGUGAUAAAUUUUACAGCCACCAAUUGGCAGUCGCAUUAAAGCCAAAUAC .((((((((.----------------------((....)).)))))))).........(((..((((((((......)))).((((.....))))....))))....)))...... ( -21.90) >DroSim_CAF1 6479 116 + 1 AUUUAUAUUAAGCAUCCACCCACAGUUUUUGAAACAUGUAAAAUGUAAAAAACAACACGGCGUAUGCGUGAUAAAUUUUACAGCCACCAAUUGGCAGUCGCAUUAAAGCCAAAUAC ........................((((((...((((.....)))).)))))).....(((..((((((((......)))).((((.....))))....))))....)))...... ( -24.20) >consensus AUUUAUAUUAAGCAUCCACCCACAGUUUUUGAAACAUGUAAAAUGUAAAAAACAACACGGCGUAUGCGUGAUAAAUUUUACAGCCACCAAUUGGCAGUCGCAUUAAAGCCAAAUAC ................................(((((.....)))))...........(((..((((((((......)))).((((.....))))....))))....)))...... (-18.04 = -18.27 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:19 2006