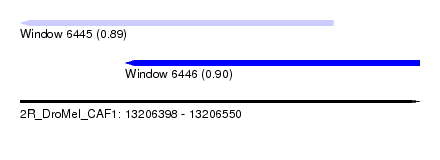
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,206,398 – 13,206,550 |
| Length | 152 |
| Max. P | 0.904120 |

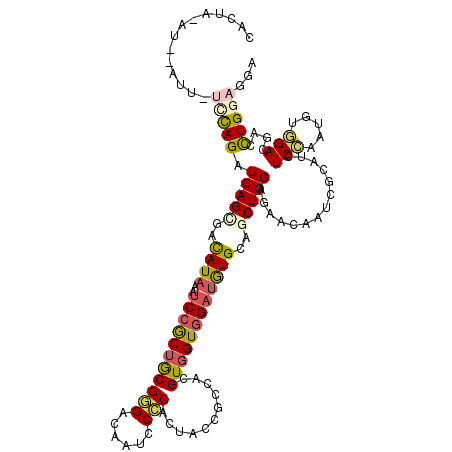
| Location | 13,206,398 – 13,206,517 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.20 |
| Mean single sequence MFE | -36.43 |
| Consensus MFE | -18.65 |
| Energy contribution | -18.85 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
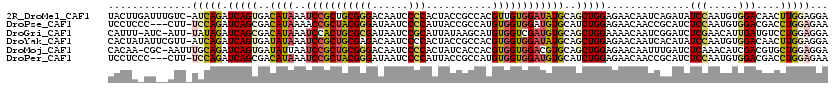
>2R_DroMel_CAF1 13206398 119 - 20766785 UACUUGAUUUGUC-AUCAGAUCAGUGACAUAAAUCCGCUGCGGGACAAUCCCCACUACCGCCACGUUGUGGAUAUGCAGCUGGAGAACAAUCAGAUAUCCAAUGUGGACAACUUGGAGGA ...((((((((..-..))))))))(((......(((((((((((......)))....((((......))))....))))).)))......)))....(((((.((.....)))))))... ( -35.20) >DroPse_CAF1 449 116 - 1 UCCUCCC---CUU-UCCAGAUCAGCGACAUAAAACCGCUACGGGAUAAUCCCCAUUACCGCCAUGUGGUGGAUGUGCAUCUGGAGAACAACCGCAUCUCCAAUGUGGACGACCUGGAGAA ..((((.---.((-(((((((..((.((((...........(((......)))....(((((....))))))))))))))))))))....((((((.....)))))).......)))).. ( -40.50) >DroGri_CAF1 7986 117 - 1 CAUUU-AUC-AUU-UAUAGAUCAGCGACAUAAAUCCACUGCGCGAUAAUCCGCAUUAUAAGCAUGUGGUCGAUGUGCAGCUGGAAAACAAUCGGAUCUCGAACAUUGAUGUCCUGGAGGA .....-...-...-......((((.(((((...(((((((((((.....((((((.......))))))....))))))).))))......(((.....)))......))))))))).... ( -32.00) >DroYak_CAF1 449 119 - 1 CACUAUAUUCGUU-AUCAGAUCAGUGAUAUAAAUCCGCUGCGAGACAAUCCCCACUACCGCCACGUGGUGGAUAUGCAGCUGGAGAACAAUCACAUAUCCAAUGUGGACAACUUGGAGGA .............-.........(((((.....((((((((((.....)).(((((((......)))))))....))))).))).....)))))...(((((.((.....)))))))... ( -33.70) >DroMoj_CAF1 1442 118 - 1 CACAA-CGC-AAUUUGCAGAUCAGUGAUAUUAAUCCGCUGCGGGACAAUCCCCACUAUCACCACGUGGUGGACGUGCAGCUGGAGAACAAUUUGAUCUCAAACAUCGACGUGCUGGAGGA (((..-((.-..((((.(((((((.........(((((((((((......))).(((((((...)))))))....))))).))).......)))))))))))...))..)))........ ( -36.69) >DroPer_CAF1 449 116 - 1 UCCUCCC---CUU-UCCAGAUCAGCGACAUAAAUCCGCUACGGGAUAAUCCCCAUUACCGCCAUGUGGUGGAUGUGCAUCUGGAGAACAACCGCAUCUCCAAUGUGGACGACCUGGAGAA ..((((.---.((-(((((((..((.((((...........(((......)))....(((((....))))))))))))))))))))....((((((.....)))))).......)))).. ( -40.50) >consensus CACUA_AU__AUU_UCCAGAUCAGCGACAUAAAUCCGCUGCGGGACAAUCCCCACUACCGCCACGUGGUGGAUGUGCAGCUGGAGAACAAUCGCAUCUCCAAUGUGGACGACCUGGAGGA ..............(((((.(((((..((((..(((((((((((......)))...........))))))))))))..)))))..............(((.....)))....)))))... (-18.65 = -18.85 + 0.20)

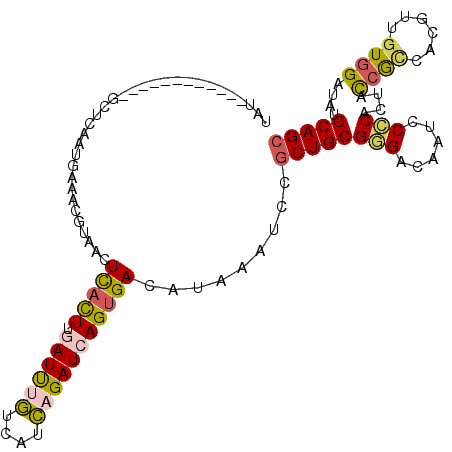
| Location | 13,206,438 – 13,206,550 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.49 |
| Mean single sequence MFE | -26.09 |
| Consensus MFE | -18.76 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.904120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13206438 112 - 20766785 UAUGAGCAUAUUAAAGCUCAAUGAAAAGUAACUUACUUGAUUUGUCAUCAGAUCAGUGACAUAAAUCCGCUGCGGGACAAUCCCCACUACCGCCACGUUGUGGAUAUGCAGC ..(((((........)))))............(((((.((((((....))))))))))).........((((((((......)))....((((......))))....))))) ( -32.20) >DroGri_CAF1 8026 96 - 1 UU---UUGU--------C---UGAUACACAAGUCAUUU-AUC-AUUUAUAGAUCAGCGACAUAAAUCCACUGCGCGAUAAUCCGCAUUAUAAGCAUGUGGUCGAUGUGCAGC ..---(((.--------(---((((....((((.....-...-))))....))))))))..........(((((((.....((((((.......))))))....))))))). ( -20.50) >DroSec_CAF1 489 100 - 1 UAU------------GCUCAAUGAAACGUAACUUACUUGAUUUGUCAUCAGAUCAGUGACUUAAAUCCGCUGCGGGACAAUCCCCACUAUCGCCACGUUGUGGAUAUGCAGC ...------------.............(((.(((((.((((((....))))))))))).))).....((((((((......))).......((((...))))....))))) ( -25.60) >DroSim_CAF1 309 100 - 1 UAU------------GCCCAAUGAAACGUAACUUACUUGAUUUGUCAUCAGAUCAGUGACUUAAAUCCGCUGCGGGACAAUCCCCACUAUCGCCACGUUGUGGAUAUGCAGC ...------------.............(((.(((((.((((((....))))))))))).))).....((((((((......))).......((((...))))....))))) ( -25.60) >DroEre_CAF1 493 100 - 1 UGU------------GCUCAAUGGAACCUAACUCACUAGAUUUGUUAACAGAUCAGUGACAUAAAUCCGCUGCGGGACAAUCCCCACUACCGUCACGUUGUGGAUAUGCAGC ...------------.......(((.......(((((.((((((....)))))))))))......)))((((((((.....)))(((.((......)).))).....))))) ( -28.72) >DroYak_CAF1 489 100 - 1 UGU------------GUUCAAUGAAACAUAACUCACUAUAUUCGUUAUCAGAUCAGUGAUAUAAAUCCGCUGCGAGACAAUCCCCACUACCGCCACGUGGUGGAUAUGCAGC (((------------(((......))))))..(((((....((.......))..))))).........(((((((.....)).(((((((......)))))))....))))) ( -23.90) >consensus UAU____________GCUCAAUGAAACGUAACUCACUUGAUUUGUCAUCAGAUCAGUGACAUAAAUCCGCUGCGGGACAAUCCCCACUACCGCCACGUUGUGGAUAUGCAGC ................................(((((.((((((....))))))))))).........((((((((......)))....((((......))))....))))) (-18.76 = -18.73 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:11 2006