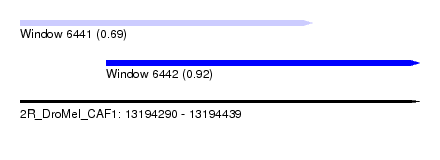
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,194,290 – 13,194,439 |
| Length | 149 |
| Max. P | 0.923905 |

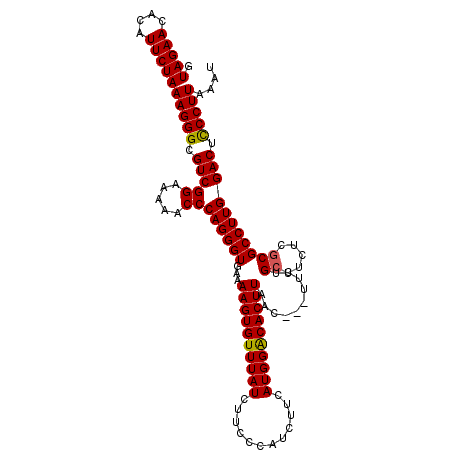
| Location | 13,194,290 – 13,194,399 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 93.99 |
| Mean single sequence MFE | -29.20 |
| Consensus MFE | -27.27 |
| Energy contribution | -27.17 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686839 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13194290 109 + 20766785 GUAGAACACAUUCUAAAGGGCGUCGGAAAAACCCAGGGUGAAAAGUGUUUAUCUUUCAAUCUUCAUGGACACUUAAC---UUCUGGUUCUCGCGCCUUGGACUCCCUUAAAU .(((((....)))))(((((.(((((.....))(((((((..((((((((((............))))))))))(((---.....)))....)))))))))).))))).... ( -30.10) >DroSec_CAF1 5102 112 + 1 GUAGAACACAUUCUAAAGGGCGUCGGAAAAACCCAGGGUGAAAAGUGUUUAUCUUCCCAUCUUCAUGGGCACUUAACUCCUUCUGCGUAUCGCGCCUUGGACUCCCUUAAAU .(((((....)))))(((((.(((((.....))((((((...(((((........(((((....))))))))))..........(((...)))))))))))).))))).... ( -30.70) >DroSim_CAF1 5099 109 + 1 GUAGAACACAUUCUAAAGGGCGUCGGAAAAACCCAGGGUGAAAAGUGUUUAUCUUCCCAUCUUCAUGGACACUUAAC---UUCUGCUUCUCGCGCCUUGGACUUCCUUAAAU .(((((....)))))(((((.(((((.....))((((((...((((((((((............))))))))))...---....((.....))))))))))).))))).... ( -26.80) >consensus GUAGAACACAUUCUAAAGGGCGUCGGAAAAACCCAGGGUGAAAAGUGUUUAUCUUCCCAUCUUCAUGGACACUUAAC___UUCUGCUUCUCGCGCCUUGGACUCCCUUAAAU .(((((....)))))(((((.(((((.....))((((((...((((((((((............))))))))))..........((.....))))))))))).))))).... (-27.27 = -27.17 + -0.11)


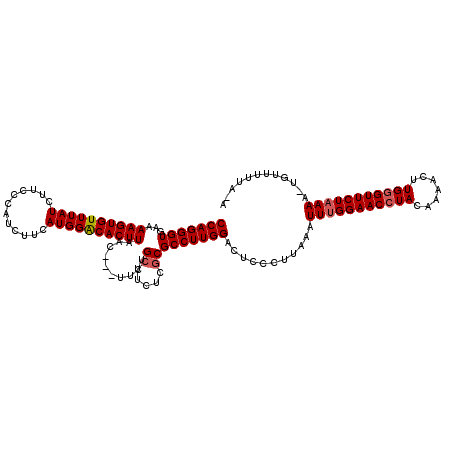
| Location | 13,194,322 – 13,194,439 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.99 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -26.02 |
| Energy contribution | -26.80 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13194322 117 + 20766785 CCAGGGUGAAAAGUGUUUAUCUUUCAAUCUUCAUGGACACUUAAC---UUCUGGUUCUCGCGCCUUGGACUCCCUUAAAUUCGGAACCUACAAAACUUGGGUUCUAAAAACAUUUUUAGA ((((((((..((((((((((............))))))))))(((---.....)))....))))))))..............((((((((.......))))))))............... ( -28.30) >DroSec_CAF1 5134 118 + 1 CCAGGGUGAAAAGUGUUUAUCUUCCCAUCUUCAUGGGCACUUAACUCCUUCUGCGUAUCGCGCCUUGGACUCCCUUAAAUUUGGAACAUACAAAACUUGGGUUCUAAAA-UGUUUUUA-A (((((((...(((((........(((((....))))))))))..........(((...))))))))))...........((((((((...((.....)).)))))))).-........-. ( -27.40) >DroSim_CAF1 5131 115 + 1 CCAGGGUGAAAAGUGUUUAUCUUCCCAUCUUCAUGGACACUUAAC---UUCUGCUUCUCGCGCCUUGGACUUCCUUAAAUUUGGAACCUACAAAACUUGGGUUCUAAAA-UGUUUUUA-A (((((((...((((((((((............))))))))))...---....((.....)))))))))...........(((((((((((.......))))))))))).-........-. ( -30.80) >consensus CCAGGGUGAAAAGUGUUUAUCUUCCCAUCUUCAUGGACACUUAAC___UUCUGCUUCUCGCGCCUUGGACUCCCUUAAAUUUGGAACCUACAAAACUUGGGUUCUAAAA_UGUUUUUA_A (((((((...((((((((((............))))))))))..........((.....)))))))))...........(((((((((((.......)))))))))))............ (-26.02 = -26.80 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:07 2006