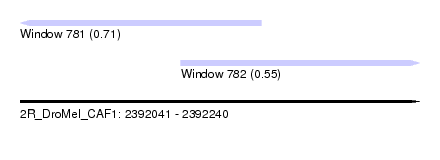
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,392,041 – 2,392,240 |
| Length | 199 |
| Max. P | 0.709827 |

| Location | 2,392,041 – 2,392,161 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.89 |
| Mean single sequence MFE | -39.42 |
| Consensus MFE | -29.45 |
| Energy contribution | -30.43 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
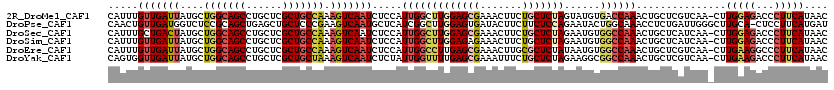
>2R_DroMel_CAF1 2392041 120 - 20766785 ACUUUGGCAGCGAGCAGGCUGCCAGCAUAAUCAACAAAUGGGUGGAGUCACAGACCAACAACCUGAUCAAGGACAUAAUCGGUCCAAGAGUUCUUACCAAGGACUCGCGGCUGUGCCUAG ...((((((((......)))))))).............((((((.((((.(.((((.....(((.....)))........))))...(((((((.....)))))))).)))).)))))). ( -40.92) >DroSec_CAF1 47910 120 - 1 ACUUUGGCAGCGAGCAGGCUGCCAGCAUAGUCAGCAAAUGGGUAGAGUCACAGACCAACAACCUGAUCAAGGACAUAAUCGGUCCAAGAGUUCUAACCAAGGACUCACGGCUGUGCCUCG ...((((((((......))))))))..((.(((.....))).))(((.(((((.((..............((((.......))))..(((((((.....)))))))..))))))).))). ( -43.30) >DroSim_CAF1 5310 120 - 1 ACUUUGGCAGCGAGCAGGCUGCCAGCAUAAUCAACAAAUGGGUAGAGUCACAGACCAACAACCUGAUCAAGGACAUAAUCGGUCCAAGAGUUCUAACCAAGGACUCGCGGCUGUGCCUCG ...((((((((......))))))))...................(((.(((((.((..............((((.......))))..(((((((.....)))))))..))))))).))). ( -41.90) >DroEre_CAF1 5744 120 - 1 ACUUUGGCAGCGAGCAGGCUGCCAGCAUAAUCAACAAAUGGGUAGCGGCCCAGACCAACAACCUAAUCAAGGACAUUAUCGGUCCAAGAGUUCUGACCAAGGACUCGCGGCUGUGCCUAG ...((((((((......)))))))).............((((((.((((((...................((((.......))))..(((((((.....)))))))).))))))))))). ( -44.00) >DroYak_CAF1 5282 120 - 1 ACUUUAGCAGCGAGCAGGCUGCCAGCAUAAUCAACCACUGGGUAGAGUCUCAGACCAACAACCUAAUCAAGGACAUUAUCGGUCCAAAUGUUCUAUCCAAGGACGCGCGGCUAUGUCUAG ....((((.(((.((.((...)).))........((..(((((((((((...)))...............((((.......))))......)))))))).)).)))...))))....... ( -28.30) >DroPer_CAF1 624 120 - 1 ACUUCGGGAGCAGCUCAGCUGCGGAGACCAUCAACAGUUGGGUGGAGGAGGAGACCAACAAGCUCAUCAAGAAUAUUGUCAGUCCGGGCGCCCUGACCAACAGCACUCGCCUGGUCCUAG .(((...((((.(((((((((..((.....))..)))))))))....(.(....)).....))))...)))..........(.(((((((..(((.....)))....))))))).).... ( -38.10) >consensus ACUUUGGCAGCGAGCAGGCUGCCAGCAUAAUCAACAAAUGGGUAGAGUCACAGACCAACAACCUGAUCAAGGACAUAAUCGGUCCAAGAGUUCUAACCAAGGACUCGCGGCUGUGCCUAG ...((((((((......)))))))).............((((((.((((.....................((((.......))))..(((((((.....)))))))..)))).)))))). (-29.45 = -30.43 + 0.98)
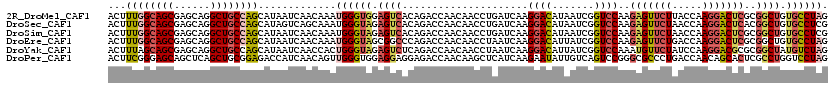
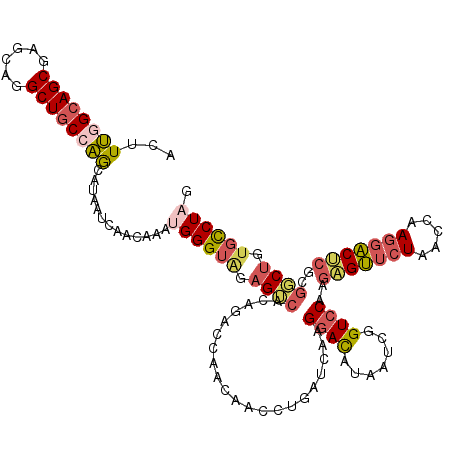
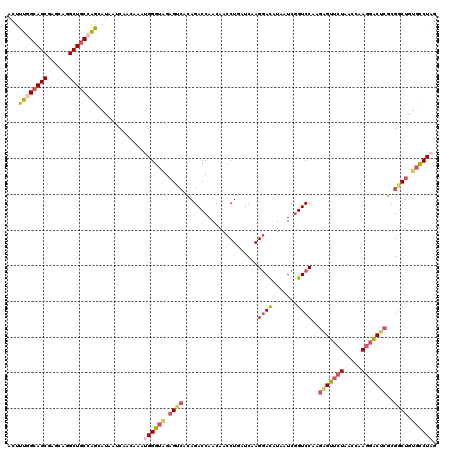
| Location | 2,392,121 – 2,392,240 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -34.82 |
| Consensus MFE | -22.78 |
| Energy contribution | -23.98 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2392121 119 + 20766785 CAUUUGUUGAUUAUGCUGGCAGCCUGCUCGCUGCCAAAGUCAAUCUCCAUUGGCUUGGAGCGAAACUUCUGCUCUAGUAUGUGACCAAACUGCUCGUCAA-CUUGGAGACCCUUCAUAAC ......((((((....(((((((......))))))).))))))(((((((((((.(((((((.......)))))))(((.((......)))))..)))))-..))))))........... ( -36.50) >DroPse_CAF1 703 119 + 1 CAACUGUUGAUGGUCUCCGCAGCUGAGCUGCUCCCGAAGUCAAUGCUCAUCGGCUUGGAGUGAUACUUCUUCUCCAGAAUACUGGCAAACCUCUGAUUGGGCUUGCA-CUCCUUCAUGAU .....((((((..((...(((((...)))))....)).))))))....((((....((((((...........((((....))))(((.(((......))).)))))-))))....)))) ( -30.00) >DroSec_CAF1 47990 119 + 1 CAUUUGCUGACUAUGCUGGCAGCCUGCUCGCUGCCAAAGUCAAUCUCCAUUGGCUUGGAGCGAAACUUCUGCUCUAGAAUGUGGCCAAACUGCUCAUCAA-CUUGGAGACCCUUCAUAAC .......(((((....(((((((......))))))).))))).(((((((((((((((((((.......)))))))......))))))............-..))))))........... ( -36.13) >DroSim_CAF1 5390 119 + 1 CAUUUGUUGAUUAUGCUGGCAGCCUGCUCGCUGCCAAAGUCAAUCUCCAUUGGCUUGGAGAGAAACUUCUGCUCUAGAAUGUGGCCAAACUGCUCAUCAA-CUUGGAGACCCUUCAUAAC .....((((((...(((((((((......))))))).............((((((((((((((....))).)))))......))))))...))..)))))-).(((((...))))).... ( -33.90) >DroEre_CAF1 5824 119 + 1 CAUUUGUUGAUUAUGCUGGCAGCCUGCUCGCUGCCAAAGUCAAUCUCCAUUGGCCUUGAGCGAAACUUGCGCUCUAUAAUGUGGCCAAACUGCUCGUCAA-CUUGAAGGCCCUUCAUAAC .....((((((...(((((((((......))))))).............((((((..(((((.......)))))........))))))...))..)))))-).(((((...))))).... ( -37.50) >DroYak_CAF1 5362 119 + 1 CAGUGGUUGAUUAUGCUGGCAGCCUGCUCGCUGCUAAAGUCAAUCUCUAUUGGUUUUGAGCGAAAUUUCUGCUCUAGAAGGCGGCCAAACUGCUCGUCAA-CUUGAAGACCCUUCAUAAC .((.((((((((....(((((((......))))))).)))))))).))...(((((((((((.......))))).....((((((......)).))))..-....))))))......... ( -34.90) >consensus CAUUUGUUGAUUAUGCUGGCAGCCUGCUCGCUGCCAAAGUCAAUCUCCAUUGGCUUGGAGCGAAACUUCUGCUCUAGAAUGUGGCCAAACUGCUCAUCAA_CUUGGAGACCCUUCAUAAC .....(((((((....(((((((......))))))).))))))).....(((((((((((((.......)))))))......))))))...............(((((...))))).... (-22.78 = -23.98 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:35 2006