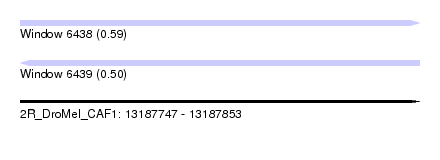
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,187,747 – 13,187,853 |
| Length | 106 |
| Max. P | 0.588618 |

| Location | 13,187,747 – 13,187,853 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -20.39 |
| Consensus MFE | -11.94 |
| Energy contribution | -13.55 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
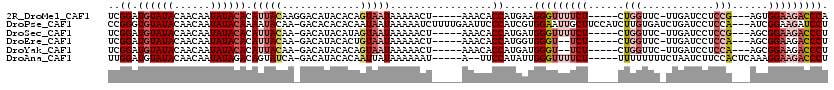
>2R_DroMel_CAF1 13187747 106 + 20766785 UCGGAUGUAUACAACAAUAUACACAUUACAAGGACAUACACAGUAAUAAAAACU-----AAACACCAUGAAGGGUUUUCU-----CUGGUUC-UUGAUCCUCCG---AGUGGAAGACCCA (((((((((((......)))))).....(((((((......(((.......)))-----((((.((.....))))))...-----...))))-)))....))))---).(((.....))) ( -21.10) >DroPse_CAF1 1832 116 + 1 CCGGGUGUAUACAACAAUAUACAAAAUACAA-GACACACACAAUAAUAAAAAUCUUUUGAAUUCCCAUCGUGGAAUUGCUUCCAUCUUGUGAUCUGAUCCUCCA---AUCGGAAGAUCCU ....((((((......))))))....(((((-((......(((.............)))((((((......)))))).......)))))))....((((.(((.---...))).)))).. ( -21.52) >DroSec_CAF1 1060 105 + 1 UCGGAUGUAUACAACAAUAUACACAUUACAA-GACAUACAUAGUAAUAAAAACU-----AAACACCAUGAUGGGUUUUCU-----CUGGUUC-UUGAUCCUCCG---AGCGGAAGACCCU ..((.((((((......)))))).(((((..-..........))))).......-----.....)).....(((((((((-----...((((-..........)---)))))))))))). ( -20.10) >DroEre_CAF1 1029 103 + 1 UCGGAUGUAUACAACAAUAUACACAUUACAA-GACAUACACUGUAAUAAAAACU-----AAACACCAUGGUGGGU--UCU-----CUGGUUC-UUGAUCCUCCA---AGCGGAAGACCCU ..((.((((((......)))))).((((((.-.........)))))).......-----.....)).....((((--..(-----(((...(-(((......))---))))))..)))). ( -21.50) >DroYak_CAF1 990 103 + 1 UCGGAUGUAUACAACAAUAUACACAUUACAA-GACAUACACAGUAAUAAAAACU-----AAACACCAUGAUGGGU--UCU-----CUGGUUC-UUGAUCCUCCA---AGCGGAAGACCCU ..((.((((((......)))))).(((((..-..........))))).......-----.....)).....((((--..(-----(((...(-(((......))---))))))..)))). ( -20.20) >DroAna_CAF1 986 107 + 1 UUGGAUGUAUACAACAAUAUAGACAGUAUCA-GACAUACACAAUUAUAAAAAAU-----A--UUCCAUAUUGGGUUUUCU-----UUUUUUUUCUAAUCUUCCACUCAAAGGAAGACCCU .(((((((((......)))))....((((..-...))))...............-----.--.))))....(((((((((-----(((...................)))))))))))). ( -17.91) >consensus UCGGAUGUAUACAACAAUAUACACAUUACAA_GACAUACACAGUAAUAAAAACU_____AAACACCAUGAUGGGUUUUCU_____CUGGUUC_UUGAUCCUCCA___AGCGGAAGACCCU ..((.((((((......)))))).(((((.............))))).................)).....(((((((((......(((............)))......))))))))). (-11.94 = -13.55 + 1.61)

| Location | 13,187,747 – 13,187,853 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.60 |
| Mean single sequence MFE | -25.78 |
| Consensus MFE | -16.80 |
| Energy contribution | -16.72 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
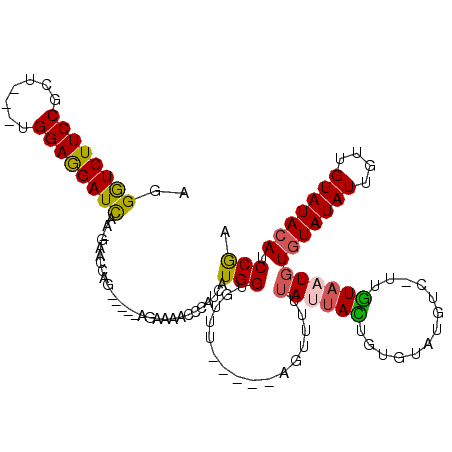
>2R_DroMel_CAF1 13187747 106 - 20766785 UGGGUCUUCCACU---CGGAGGAUCAA-GAACCAG-----AGAAAACCCUUCAUGGUGUUU-----AGUUUUUAUUACUGUGUAUGUCCUUGUAAUGUGUAUAUUGUUGUAUACAUCCGA .(((((((((...---.)))))))(((-(.((((.-----.(((.....))).))))...(-----(((.......))))........))))..(((((((((....))))))))))).. ( -24.00) >DroPse_CAF1 1832 116 - 1 AGGAUCUUCCGAU---UGGAGGAUCAGAUCACAAGAUGGAAGCAAUUCCACGAUGGGAAUUCAAAAGAUUUUUAUUAUUGUGUGUGUC-UUGUAUUUUGUAUAUUGUUGUAUACACCCGG ..((((((((...---.)))))))).....(((((((((((....))))..((((((((((.....)))))))))).........)))-))))....(((((((....)))))))..... ( -33.10) >DroSec_CAF1 1060 105 - 1 AGGGUCUUCCGCU---CGGAGGAUCAA-GAACCAG-----AGAAAACCCAUCAUGGUGUUU-----AGUUUUUAUUACUAUGUAUGUC-UUGUAAUGUGUAUAUUGUUGUAUACAUCCGA ..((((((((...---.))))))))..-......(-----(((.....((((((((((...-----.........))))))).)))))-))...(((((((((....))))))))).... ( -26.20) >DroEre_CAF1 1029 103 - 1 AGGGUCUUCCGCU---UGGAGGAUCAA-GAACCAG-----AGA--ACCCACCAUGGUGUUU-----AGUUUUUAUUACAGUGUAUGUC-UUGUAAUGUGUAUAUUGUUGUAUACAUCCGA ..((((((((...---.))))))))((-((((.((-----(..--(((......))).)))-----.))))))(((((((........-)))))))((((((((....)))))))).... ( -25.80) >DroYak_CAF1 990 103 - 1 AGGGUCUUCCGCU---UGGAGGAUCAA-GAACCAG-----AGA--ACCCAUCAUGGUGUUU-----AGUUUUUAUUACUGUGUAUGUC-UUGUAAUGUGUAUAUUGUUGUAUACAUCCGA ..((((((((...---.))))))))..-......(-----(((--...((((((((((...-----.........))))))).)))))-))...(((((((((....))))))))).... ( -25.10) >DroAna_CAF1 986 107 - 1 AGGGUCUUCCUUUGAGUGGAAGAUUAGAAAAAAAA-----AGAAAACCCAAUAUGGAA--U-----AUUUUUUAUAAUUGUGUAUGUC-UGAUACUGUCUAUAUUGUUGUAUACAUCCAA .(((((((((.......)))))))...........-----........(((((((((.--(-----(((...((((......))))..-.))))...)))))))))..........)).. ( -20.50) >consensus AGGGUCUUCCGCU___UGGAGGAUCAA_GAACCAG_____AGAAAACCCAUCAUGGUGUUU_____AGUUUUUAUUACUGUGUAUGUC_UUGUAAUGUGUAUAUUGUUGUAUACAUCCGA ..((((((((.......))))))))............................(((................((((((.............))))))(((((((....))))))).))). (-16.80 = -16.72 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:05 2006