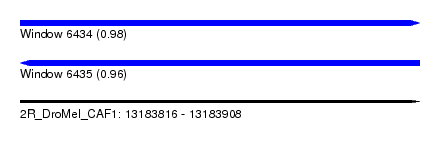
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,183,816 – 13,183,908 |
| Length | 92 |
| Max. P | 0.984189 |

| Location | 13,183,816 – 13,183,908 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 76.68 |
| Mean single sequence MFE | -39.28 |
| Consensus MFE | -29.14 |
| Energy contribution | -28.32 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.41 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13183816 92 + 20766785 UUAA-AAUGGAAUGGCCAUGGACAACACGUUGGGUCACAUGCUUUCAGGCCUCCGAUUC-----GGUUUCAGGC-----UGGGUCGGAGGUCUGUUUUUAUCA-- ....-.(((((((((((...(((.....))).)))))........((((((((((((((-----(((.....))-----))))))))))))))).))))))..-- ( -36.40) >DroPse_CAF1 80454 104 + 1 UUAA-AAUGGAAUGGUCAUGGACAACUCUUGGUGUUAGCUGCAGCCAGGCCUCCGACUCAGCUCAGUUCCCGACAGAGCAGAGUCGGAGGUCUGCUUUAAUCCGC ....-...(((...((....((((.(....).))))....))(((.(((((((((((((.((((.(((...))).)))).))))))))))))))))....))).. ( -41.30) >DroEre_CAF1 78931 94 + 1 UUAA-AAUGGAAUGGCCAUGGACAACACGUUGGGUCAAAUGCUGUCAGGCCUCCGAUUC-----GGUUUCCGAC-----UGGGUCGGAGGUCUAUUUUUAUCAUU ....-(((((..(((((...(((.....))).))))).........(((((((((((((-----((((...)))-----)))))))))))))).......))))) ( -35.20) >DroYak_CAF1 79909 94 + 1 UUAA-AAUGGAAUGGCCAUGGACAACAAGUUGGGUCAAAUGCUGUCAGGCCUCCGAUUC-----GGUUCCCGAC-----UGGGUCGGAGGUCUGUUUUUAUCAUC ....-.((((..(((((...(((.....))).)))))........((((((((((((((-----((((...)))-----)))))))))))))))......)))). ( -36.70) >DroAna_CAF1 76557 95 + 1 UUAAAAAUGGGAAGGCCAUGGACAAUUGCAGGGGUUACGUGUAGUCAGACCUCCGAUUU-----GGUUCCUGAC-----UGGAUCGGAGGUCUGGUUCCAUUAUC .....(((((((............((((((.(.....).))))))(((((((((((((.-----.(((...)))-----..))))))))))))).)))))))... ( -41.20) >DroPer_CAF1 74519 104 + 1 UUAA-AAUGGAAUGGUCAUGGACAACUCUUGGUGUUAGCUGCAGCCAGGCCUCCGACUCAGUUCAGUUCCCGACAGAGCUGAGUCGGAGGUCUGCUUUAAUCCAC ....-..((((...((....((((.(....).))))....))(((.((((((((((((((((((.(((...))).)))))))))))))))))))))....)))). ( -44.90) >consensus UUAA_AAUGGAAUGGCCAUGGACAACACGUGGGGUCAAAUGCAGUCAGGCCUCCGAUUC_____GGUUCCCGAC_____UGGGUCGGAGGUCUGUUUUUAUCAUC ......((((((..((....(((..........)))....))...((((((((((((((.((((.(((...))).)))).)))))))))))))).)))))).... (-29.14 = -28.32 + -0.83)


| Location | 13,183,816 – 13,183,908 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 76.68 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -14.10 |
| Energy contribution | -15.13 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13183816 92 - 20766785 --UGAUAAAAACAGACCUCCGACCCA-----GCCUGAAACC-----GAAUCGGAGGCCUGAAAGCAUGUGACCCAACGUGUUGUCCAUGGCCAUUCCAUU-UUAA --.(((.....(((.(((((((..(.-----..........-----)..))))))).)))..(((((((......)))))))))).((((.....)))).-.... ( -23.00) >DroPse_CAF1 80454 104 - 1 GCGGAUUAAAGCAGACCUCCGACUCUGCUCUGUCGGGAACUGAGCUGAGUCGGAGGCCUGGCUGCAGCUAACACCAAGAGUUGUCCAUGACCAUUCCAUU-UUAA ..(((....(((((.((((((((((.((((.((.....)).)))).)))))))))).)).)))((((((.........))))))..........)))...-.... ( -39.70) >DroEre_CAF1 78931 94 - 1 AAUGAUAAAAAUAGACCUCCGACCCA-----GUCGGAAACC-----GAAUCGGAGGCCUGACAGCAUUUGACCCAACGUGUUGUCCAUGGCCAUUCCAUU-UUAA .....(((((.......((((((...-----))))))....-----.....(((((((.((((((((.(......).))))))))...))))..))).))-))). ( -28.00) >DroYak_CAF1 79909 94 - 1 GAUGAUAAAAACAGACCUCCGACCCA-----GUCGGGAACC-----GAAUCGGAGGCCUGACAGCAUUUGACCCAACUUGUUGUCCAUGGCCAUUCCAUU-UUAA ((((.............(((((....-----.((((...))-----)).)))))((((.(((((((..(......)..)))))))...))))....))))-.... ( -26.30) >DroAna_CAF1 76557 95 - 1 GAUAAUGGAACCAGACCUCCGAUCCA-----GUCAGGAACC-----AAAUCGGAGGUCUGACUACACGUAACCCCUGCAAUUGUCCAUGGCCUUCCCAUUUUUAA ....(((((..((((((((((((((.-----....)))...-----...))))))))))).......(((.....))).....)))))((.....))........ ( -28.10) >DroPer_CAF1 74519 104 - 1 GUGGAUUAAAGCAGACCUCCGACUCAGCUCUGUCGGGAACUGAACUGAGUCGGAGGCCUGGCUGCAGCUAACACCAAGAGUUGUCCAUGACCAUUCCAUU-UUAA (((((....(((((.((((((((((((.((.((.....)).)).)))))))))))).)).)))((((((.........))))))..........))))).-.... ( -38.80) >consensus GAUGAUAAAAACAGACCUCCGACCCA_____GUCGGGAACC_____GAAUCGGAGGCCUGACAGCAUCUAACCCAACGAGUUGUCCAUGGCCAUUCCAUU_UUAA ...........(((.(((((((..(.((((.((.....)).)))).)..))))))).)))............................................. (-14.10 = -15.13 + 1.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:09:01 2006