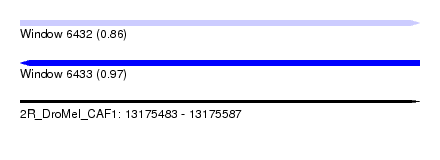
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,175,483 – 13,175,587 |
| Length | 104 |
| Max. P | 0.968420 |

| Location | 13,175,483 – 13,175,587 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -25.82 |
| Energy contribution | -25.70 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
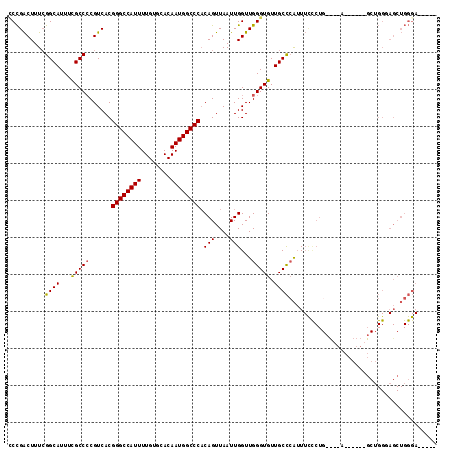
>2R_DroMel_CAF1 13175483 104 + 20766785 UCCGACUUUCGGCAUUUCGCCCCGUCACGGGCCAUUUUGUGCACAAUGGCCCACAGUUAAUUGGUUGGGUGUUGCCCAUUUCUCUGCU--A------GCUGGUAGCUGGGAA---- .(((.....))).......(((.(..((((((((((........)))))))).((((((...((.((((.....))))...))....)--)------))))))..).)))..---- ( -36.10) >DroSec_CAF1 69553 97 + 1 CCCGACUUUAGGCAUUUCGCCGCGUCACGGGCCAUUUUGUGCACAAUGGCCCACAGUUAAUUGGUUGGGUGUUGCCCAUUUCUCU---------------GGGAGCUGGGAU---- (((((((...(((.....))).......((((((((........))))))))..........))))))).....((((.(((...---------------..))).))))..---- ( -35.30) >DroEre_CAF1 70664 111 + 1 GCCGACUUUCGGCAUUUCGCCCCGUCACGGGCCAUUUUGUGCACAAUGGCCCACAGUUAAUUGGUUGGGUGUUGCCCAUUUCCCUGGCAUACUGGGUGCUGGGAGCUGGGA----- ((((.....))))....(((((......((((((((........)))))))).(((....)))...)))))...((((.(((((.(((((.....)))))))))).)))).----- ( -47.30) >DroYak_CAF1 71608 114 + 1 CCCGACUUUCGGCAUUUCGCCCCGUCACGGGCCAUUUUGUGCACAAUGGCCCACAGUUAAUUGGUUGGGUGUUGCCCAUUUUCCUGCC--ACUGGGCGCUGGUAGCUGGGAUCCUG ((((.((.(((((....(((((......((((((((........)))))))).(((....)))...)))))..(((((..........--..)))))))))).)).))))...... ( -42.10) >DroAna_CAF1 68864 80 + 1 CCUGGCUUUCGGCAUUUUGCCCCGUCACGGGCCAUUUUGUGCACAAUGGCCCCCAGUUAAUUGGUUGGGUGUUGCUGGGU------------------------------------ ((..((....(((.....)))....(((((((((((........))))))))((((........)))))))..))..)).------------------------------------ ( -29.60) >consensus CCCGACUUUCGGCAUUUCGCCCCGUCACGGGCCAUUUUGUGCACAAUGGCCCACAGUUAAUUGGUUGGGUGUUGCCCAUUUCCCUG____A______GCUGGGAGCUGGGA_____ ..........((((...(((((......((((((((........)))))))).(((....)))...))))).))))........................................ (-25.82 = -25.70 + -0.12)

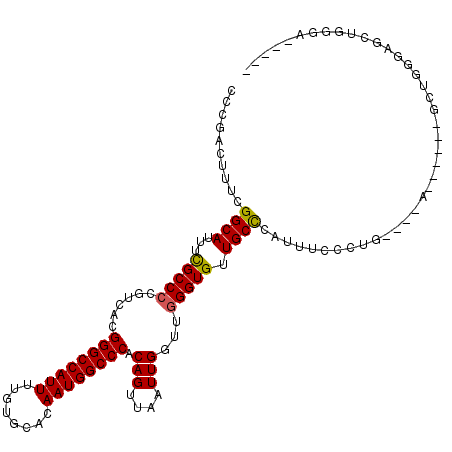
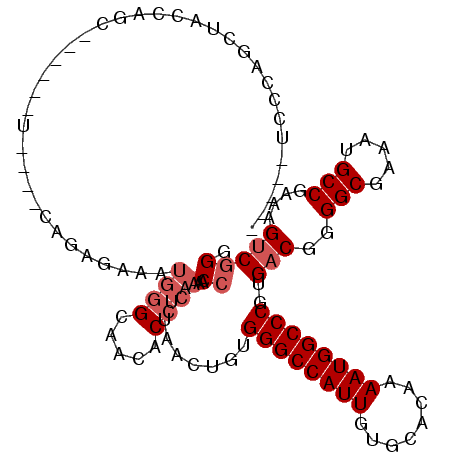
| Location | 13,175,483 – 13,175,587 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 78.42 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -23.68 |
| Energy contribution | -24.68 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13175483 104 - 20766785 ----UUCCCAGCUACCAGC------U--AGCAGAGAAAUGGGCAACACCCAACCAAUUAACUGUGGGCCAUUGUGCACAAAAUGGCCCGUGACGGGGCGAAAUGCCGAAAGUCGGA ----.(((((((.....))------)--.((((..((.(((...........))).))..))))((((((((........)))))))).....)))).......(((.....))). ( -32.30) >DroSec_CAF1 69553 97 - 1 ----AUCCCAGCUCCC---------------AGAGAAAUGGGCAACACCCAACCAAUUAACUGUGGGCCAUUGUGCACAAAAUGGCCCGUGACGCGGCGAAAUGCCUAAAGUCGGG ----..((((.(((..---------------.)))...)))).....(((..............((((((((........))))))))..(((..(((.....)))....)))))) ( -32.80) >DroEre_CAF1 70664 111 - 1 -----UCCCAGCUCCCAGCACCCAGUAUGCCAGGGAAAUGGGCAACACCCAACCAAUUAACUGUGGGCCAUUGUGCACAAAAUGGCCCGUGACGGGGCGAAAUGCCGAAAGUCGGC -----.....(((((...(((.((((......((....((((.....)))).)).....)))).((((((((........)))))))))))..))))).....((((.....)))) ( -40.40) >DroYak_CAF1 71608 114 - 1 CAGGAUCCCAGCUACCAGCGCCCAGU--GGCAGGAAAAUGGGCAACACCCAACCAAUUAACUGUGGGCCAUUGUGCACAAAAUGGCCCGUGACGGGGCGAAAUGCCGAAAGUCGGG ......(((.((......(((((.((--.((.((....((((.....)))).))..........((((((((........)))))))))).)).)))))....))(....)..))) ( -42.10) >DroAna_CAF1 68864 80 - 1 ------------------------------------ACCCAGCAACACCCAACCAAUUAACUGGGGGCCAUUGUGCACAAAAUGGCCCGUGACGGGGCAAAAUGCCGAAAGCCAGG ------------------------------------.((..((..(((....(((......)))((((((((........)))))))))))....(((.....)))....))..)) ( -27.60) >consensus _____UCCCAGCUACCAGC______U____CAGAGAAAUGGGCAACACCCAACCAAUUAACUGUGGGCCAUUGUGCACAAAAUGGCCCGUGACGGGGCGAAAUGCCGAAAGUCGGG ......................................((((.....)))).((..........((((((((........))))))))..(((..(((.....)))....))))). (-23.68 = -24.68 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:59 2006