| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,144,673 – 13,144,813 |
| Length | 140 |
| Max. P | 0.875665 |

| Location | 13,144,673 – 13,144,783 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.19 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -9.47 |
| Energy contribution | -10.00 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
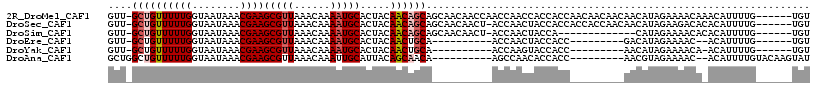
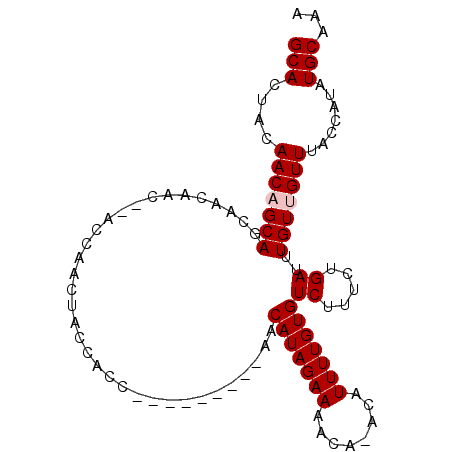
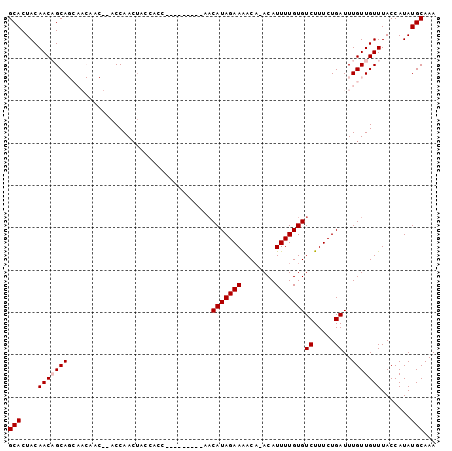
>2R_DroMel_CAF1 13144673 110 + 20766785 GUU-GCUGUUUUUGGUAAUAAACGAAGCGUUAAACAAAAUGCACUACAACAGCAGCAACAACCAACCAACCACCACCAACAACAACAACAUAGAAAACAAACAUUUUG------UGU (((-((((((.(((((.....))...(((((......)))))....))).)))))))))............................((((((((........)))))------))) ( -23.00) >DroSec_CAF1 38468 109 + 1 GUU-GCUGUUUUUGGUAAUAAACGAAGCGUUAAACAAAAUGCACUACAACAGCAGCAACAACU-ACCAACUACCACCACCACCAACAACAUAGAAGACACACAUUUUG------UGU (((-((((((.(((((.....))...(((((......)))))....))).)))))))))....-.......................((((((((........)))))------))) ( -23.80) >DroSim_CAF1 40272 96 + 1 GUU-GCUGUUUUUGGUAAUAAACGAAGCGUUAAACAAAAUGCACUACAACAGCAGCAACAACU-ACCAACUACCA-------------CAUAGAAAACACACAUUUUG------UGU (((-((((((.(((((.....))...(((((......)))))....))).)))))))))....-..........(-------------(((((((........)))))------))) ( -23.00) >DroEre_CAF1 39062 89 + 1 GUU-GCUGUUUUUGGUAAUAAACGAAGCGUUAAACAAAAUGCACUACAACUGCA----------ACCAACUACCACC---------GACAUAGAAAAC--ACAUUUUG------UGU (((-((.(((((((........))))(((((......))))).....))).)))----------))...........---------.((((((((...--...)))))------))) ( -16.70) >DroYak_CAF1 39368 90 + 1 GUU-GCUGUUUUUGGUAAUAAACGAAGCGUUAAACAAAAUGCACUACAACUGCA----------ACCAAGUACCACC---------AACAUAGAAAACA-ACAUUUUG------UGU (((-((.(((((((........))))(((((......))))).....))).)))----------))...........---------.((((((((....-...)))))------))) ( -16.10) >DroAna_CAF1 38627 96 + 1 GCUGGCUGUUUUUGGUAAUAAACGAAGCGUUAAACAAAUUGCAUUACAGCAACA----------AGCCAACACCACC---------AACGUAGAAAAC--ACAUUUUGUACAAGUAU (((.(((((.((((........))))(((..........)))...)))))....----------)))..........---------...(((.((((.--...)))).)))...... ( -14.80) >consensus GUU_GCUGUUUUUGGUAAUAAACGAAGCGUUAAACAAAAUGCACUACAACAGCA__________ACCAACUACCACC_________AACAUAGAAAACA_ACAUUUUG______UGU ....((((((((((........))))(((((......))))).....))))))................................................................ ( -9.47 = -10.00 + 0.53)
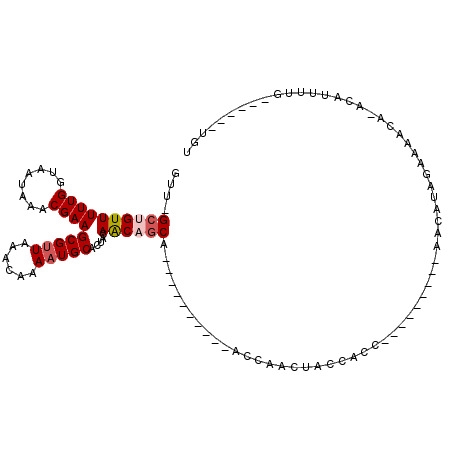
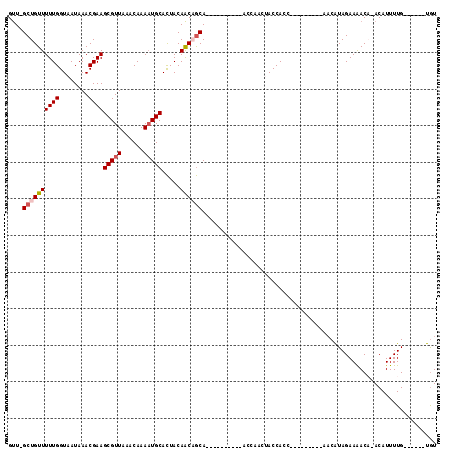
| Location | 13,144,712 – 13,144,813 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 82.94 |
| Mean single sequence MFE | -14.23 |
| Consensus MFE | -8.46 |
| Energy contribution | -8.86 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13144712 101 + 20766785 GCACUACAACAGCAGCAACAACCAACCAACCACCACCAACAACAACAACAUAGAAAACAAACAUUUUGUGUCUUUCUGAUUUGUUGUUUACCAUAUGCAAA (((.......((((((((..............................(((((((........)))))))((.....)).)))))))).......)))... ( -12.04) >DroSec_CAF1 38507 100 + 1 GCACUACAACAGCAGCAACAACU-ACCAACUACCACCACCACCAACAACAUAGAAGACACACAUUUUGUGUCUUUCUGAUUUGUUGUUUACCAUAUGCAAA (((..........((......))-...................((((((((((((((((((.....)))))).)))))...))))))).......)))... ( -17.60) >DroSim_CAF1 40311 87 + 1 GCACUACAACAGCAGCAACAACU-ACCAACUACCA-------------CAUAGAAAACACACAUUUUGUGUCUUUCUGAUUUGUUGUUUACCAUAUGCAAA (((.......((((((((.....-...........-------------..(((((((((((.....))))).))))))..)))))))).......)))... ( -14.19) >DroEre_CAF1 39101 80 + 1 GCACUACAACUGCA----------ACCAACUACCACC---------GACAUAGAAAAC--ACAUUUUGUGUCUUUCUGAUUUGUUGUUUACCAUAUGCAAA (((........(((----------((...........---------(((((((((...--...)))))))))..........)))))........)))... ( -14.19) >DroYak_CAF1 39407 81 + 1 GCACUACAACUGCA----------ACCAAGUACCACC---------AACAUAGAAAACA-ACAUUUUGUGUCUUUCUGAUUUGUUGUUUACCAUAUGCAAA (((........(((----------((.((((......---------.((((((((....-...)))))))).......)))))))))........)))... ( -13.13) >consensus GCACUACAACAGCAGCAACAAC__ACCAACUACCACC_________AACAUAGAAAACA_ACAUUUUGUGUCUUUCUGAUUUGUUGUUUACCAUAUGCAAA (((....(((((((..................................(((((((........)))))))((.....))..))))))).......)))... ( -8.46 = -8.86 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:55 2006