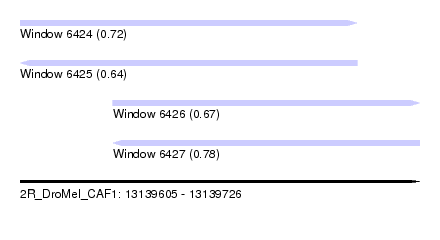
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,139,605 – 13,139,726 |
| Length | 121 |
| Max. P | 0.777502 |

| Location | 13,139,605 – 13,139,707 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -25.19 |
| Consensus MFE | -17.37 |
| Energy contribution | -18.07 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
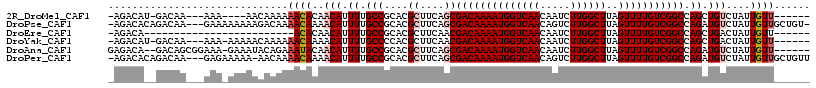
>2R_DroMel_CAF1 13139605 102 + 20766785 -AGACAU-GACAA---AAA----AACAAAAAACACAACAUUUUGCCGCACGCUUCAGCGACAAAAUGGUCAACAAUCUUGGCUUAGUUUUGUCGGCCAGCUGUCUAUUGUU------ -((((..-(((((---((.----..............(((((((.(((........))).)))))))(((((.....)))))....)))))))((....))))))......------ ( -24.30) >DroPse_CAF1 33321 112 + 1 -AGACACAGACAA---GAAAAAAAAGACAAAACAAAACAUUUUGCCGCACGCUUCAGCGACAAAAUGGUCAACAGUCUUGGCUUAGUUUUGUCGGCCAGAUGUCUAUUGUUGCUGU- -....((((.(((---.((......((((((((....(((((((.(((........))).)))))))(((((.....)))))...))))))))(((.....)))..)).)))))))- ( -31.50) >DroEre_CAF1 33925 85 + 1 -AGACA-------------------------ACACAACAUUUUGCCGCACGCUUCAACGACAAAAUGGUCAACAAUCUUGGCUUAGUUUUGUCGGCCAGCUGACUAUUGUU------ -.....-------------------------....((((........((.(((....(((((((((((((((.....))))))..)))))))))...))))).....))))------ ( -18.52) >DroYak_CAF1 34096 105 + 1 -AGACAU-GACAA---AAA-AAAAACAAAAAACAAAACAUUUUGCCCCACGCUUCAACGACAAAAUGGUCAACAAUCUUGGCUUAGUUUUGUCGGCCAGCUGACUAUUGUU------ -......-(((((---...-.....(((((.........)))))...((.(((....(((((((((((((((.....))))))..)))))))))...)))))....)))))------ ( -19.90) >DroAna_CAF1 33440 108 + 1 GAGACA--GACAGCGGAAA-GAAAUACAGAAAUACAACAUUUUGCCGCACGCUUCAGCGACAAAAUGGUCAACAAUCUUGGCUUAGUUUUGUCGGCCAGAUGUCUAUUGUU------ .(((((--....((((...-(((((.............))))).)))).........(((((((((((((((.....))))))..)))))))))......)))))......------ ( -27.52) >DroPer_CAF1 33033 112 + 1 -AGACACAGACAA---GAGAAAAA-AACAAAACAAAACAUUUUGCCGCACGCUUCAGCGACAAAAUGGUCAACAGUCUUGGCUUAGUUUUGUCGGCCAGAUGUCUAUUGUUGCUGUU -............---........-((((.(((((.((((((.(((....((....))((((((((((((((.....))))))..))))))))))).))))))...)))))..)))) ( -29.40) >consensus _AGACA__GACAA___AAA_AAAAAAAAAAAACAAAACAUUUUGCCGCACGCUUCAGCGACAAAAUGGUCAACAAUCUUGGCUUAGUUUUGUCGGCCAGAUGUCUAUUGUU______ ..............................((((..(((.((.(((....((....))((((((((((((((.....))))))..))))))))))).)).)))....))))...... (-17.37 = -18.07 + 0.70)

| Location | 13,139,605 – 13,139,707 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -20.66 |
| Energy contribution | -20.38 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
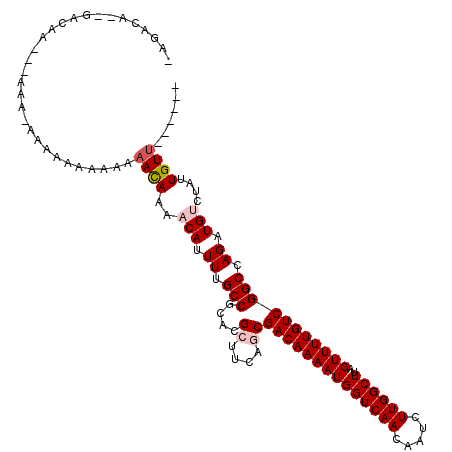
>2R_DroMel_CAF1 13139605 102 - 20766785 ------AACAAUAGACAGCUGGCCGACAAAACUAAGCCAAGAUUGUUGACCAUUUUGUCGCUGAAGCGUGCGGCAAAAUGUUGUGUUUUUUGUU----UUU---UUGUC-AUGUCU- ------(((((.(.(((((.((.((((((..((......)).)))))).))((((((((((........))))))))))))))).)...)))))----...---.....-......- ( -26.50) >DroPse_CAF1 33321 112 - 1 -ACAGCAACAAUAGACAUCUGGCCGACAAAACUAAGCCAAGACUGUUGACCAUUUUGUCGCUGAAGCGUGCGGCAAAAUGUUUUGUUUUGUCUUUUUUUUC---UUGUCUGUGUCU- -...........((((((..(((.((.((((..(((.((((((....((.(((((((((((........))))))))))).)).)))))).)))..)))).---))))).))))))- ( -31.60) >DroEre_CAF1 33925 85 - 1 ------AACAAUAGUCAGCUGGCCGACAAAACUAAGCCAAGAUUGUUGACCAUUUUGUCGUUGAAGCGUGCGGCAAAAUGUUGUGU-------------------------UGUCU- ------.((((((((((((((((............)))).....))))))(((((((((((........)))))))))))...)))-------------------------)))..- ( -25.80) >DroYak_CAF1 34096 105 - 1 ------AACAAUAGUCAGCUGGCCGACAAAACUAAGCCAAGAUUGUUGACCAUUUUGUCGUUGAAGCGUGGGGCAAAAUGUUUUGUUUUUUGUUUUU-UUU---UUGUC-AUGUCU- ------.(((...(((....))).(((((((..(((.(((((.....((.(((((((((.(........).))))))))).))....))))).))).-.))---)))))-.)))..- ( -22.00) >DroAna_CAF1 33440 108 - 1 ------AACAAUAGACAUCUGGCCGACAAAACUAAGCCAAGAUUGUUGACCAUUUUGUCGCUGAAGCGUGCGGCAAAAUGUUGUAUUUCUGUAUUUC-UUUCCGCUGUC--UGUCUC ------....(((((((.(((((............))))(((....((((.((((((((((........))))))))))))))....))).......-.....).))))--)))... ( -26.70) >DroPer_CAF1 33033 112 - 1 AACAGCAACAAUAGACAUCUGGCCGACAAAACUAAGCCAAGACUGUUGACCAUUUUGUCGCUGAAGCGUGCGGCAAAAUGUUUUGUUUUGUU-UUUUUCUC---UUGUCUGUGUCU- .(((((((((((((...((((((............))).)))))))))..(((((((((((........)))))))))))............-........---))).))))....- ( -29.30) >consensus ______AACAAUAGACAGCUGGCCGACAAAACUAAGCCAAGAUUGUUGACCAUUUUGUCGCUGAAGCGUGCGGCAAAAUGUUGUGUUUUUUGUUUUU_UUU___UUGUC__UGUCU_ ........((((((.(...((((............)))).).))))))..(((((((((((........)))))))))))..................................... (-20.66 = -20.38 + -0.28)
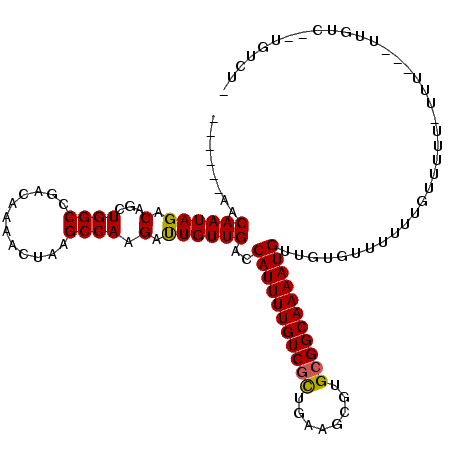
| Location | 13,139,633 – 13,139,726 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.27 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -20.28 |
| Energy contribution | -20.39 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
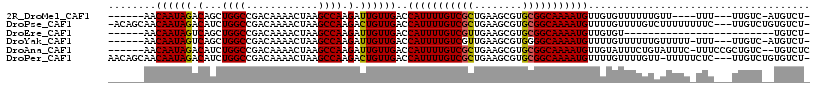
>2R_DroMel_CAF1 13139633 93 + 20766785 CAUUUUGCCGCACGCUUCAGCGACAAAAUGGUCAACAAUCUUGGCUUAGUUUUGUCGGCCAGCUGUCUAUUGUU---------------------------GUUGUAAUCACAAUGGGUA ......(((....((....))((((((((((((((.....))))))..)))))))))))......((((((((.---------------------------((....)).)))))))).. ( -24.70) >DroPse_CAF1 33354 114 + 1 CAUUUUGCCGCACGCUUCAGCGACAAAAUGGUCAACAGUCUUGGCUUAGUUUUGUCGGCCAGAUGUCUAUUGUUGCUGU------UGCUGUUGCCGUUGCUGUUGUAAUUACAAUGGCUA ......(((((((((..((((((((.((..((..(((.(((.((((..........))))))))))..))..))..)))------)))))..).)).))).(((((....)))))))).. ( -35.80) >DroEre_CAF1 33936 93 + 1 CAUUUUGCCGCACGCUUCAACGACAAAAUGGUCAACAAUCUUGGCUUAGUUUUGUCGGCCAGCUGACUAUUGUU---------------------------GUUGUAAUUACAAUAGGUA .....((((.((.(((....(((((((((((((((.....))))))..)))))))))...)))))........(---------------------------(((((....)))))))))) ( -24.80) >DroYak_CAF1 34127 93 + 1 CAUUUUGCCCCACGCUUCAACGACAAAAUGGUCAACAAUCUUGGCUUAGUUUUGUCGGCCAGCUGACUAUUGUU---------------------------GUUGUAAUUACAAUAGCUA ..........((.(((....(((((((((((((((.....))))))..)))))))))...)))))......(((---------------------------(((((....)))))))).. ( -24.90) >DroAna_CAF1 33474 90 + 1 CAUUUUGCCGCACGCUUCAGCGACAAAAUGGUCAACAAUCUUGGCUUAGUUUUGUCGGCCAGAUGUCUAUUGUU---------------------------GUUGUAAUUACAAUGG--- (((((.(((....((....))((((((((((((((.....))))))..))))))))))).))))).(((((((.---------------------------((....)).)))))))--- ( -26.80) >DroPer_CAF1 33065 120 + 1 CAUUUUGCCGCACGCUUCAGCGACAAAAUGGUCAACAGUCUUGGCUUAGUUUUGUCGGCCAGAUGUCUAUUGUUGCUGUUGCUGUUGCUGUUGCCGUUGCUGUUGUAAUUACAAUGGCUA ......((.(((.((..((((((((.((..((..(((.(((.((((..........))))))))))..))..))..))))))))..))))).))....((((((((....)))))))).. ( -38.80) >consensus CAUUUUGCCGCACGCUUCAGCGACAAAAUGGUCAACAAUCUUGGCUUAGUUUUGUCGGCCAGAUGUCUAUUGUU___________________________GUUGUAAUUACAAUGGCUA ......(((....((....))((((((((((((((.....))))))..))))))))))).......(((((((.....................................)))))))... (-20.28 = -20.39 + 0.11)

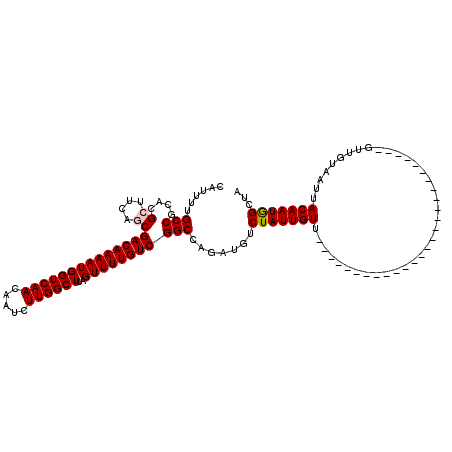

| Location | 13,139,633 – 13,139,726 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.27 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -20.41 |
| Energy contribution | -20.13 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13139633 93 - 20766785 UACCCAUUGUGAUUACAAC---------------------------AACAAUAGACAGCUGGCCGACAAAACUAAGCCAAGAUUGUUGACCAUUUUGUCGCUGAAGCGUGCGGCAAAAUG ......((((....))))(---------------------------((((((.......((((............))))..)))))))..(((((((((((........))))))))))) ( -23.40) >DroPse_CAF1 33354 114 - 1 UAGCCAUUGUAAUUACAACAGCAACGGCAACAGCA------ACAGCAACAAUAGACAUCUGGCCGACAAAACUAAGCCAAGACUGUUGACCAUUUUGUCGCUGAAGCGUGCGGCAAAAUG ..(((.((((..........)))).)))....((.------...))..((((((...((((((............))).)))))))))..(((((((((((........))))))))))) ( -31.90) >DroEre_CAF1 33936 93 - 1 UACCUAUUGUAAUUACAAC---------------------------AACAAUAGUCAGCUGGCCGACAAAACUAAGCCAAGAUUGUUGACCAUUUUGUCGUUGAAGCGUGCGGCAAAAUG ......((((....)))).---------------------------..((((((((...((((............)))).))))))))..(((((((((((........))))))))))) ( -23.90) >DroYak_CAF1 34127 93 - 1 UAGCUAUUGUAAUUACAAC---------------------------AACAAUAGUCAGCUGGCCGACAAAACUAAGCCAAGAUUGUUGACCAUUUUGUCGUUGAAGCGUGGGGCAAAAUG ......((((....)))).---------------------------..((((((((...((((............)))).))))))))..(((((((((.(........).))))))))) ( -20.00) >DroAna_CAF1 33474 90 - 1 ---CCAUUGUAAUUACAAC---------------------------AACAAUAGACAUCUGGCCGACAAAACUAAGCCAAGAUUGUUGACCAUUUUGUCGCUGAAGCGUGCGGCAAAAUG ---...((((....))))(---------------------------((((((.......((((............))))..)))))))..(((((((((((........))))))))))) ( -23.70) >DroPer_CAF1 33065 120 - 1 UAGCCAUUGUAAUUACAACAGCAACGGCAACAGCAACAGCAACAGCAACAAUAGACAUCUGGCCGACAAAACUAAGCCAAGACUGUUGACCAUUUUGUCGCUGAAGCGUGCGGCAAAAUG ..(((.((((....))))..(((...((..((((....((....)).......((((..(((.(((((...((......))..))))).)))...))))))))..)).))))))...... ( -32.40) >consensus UAGCCAUUGUAAUUACAAC___________________________AACAAUAGACAGCUGGCCGACAAAACUAAGCCAAGAUUGUUGACCAUUUUGUCGCUGAAGCGUGCGGCAAAAUG ......((((....))))..............................((((((.(...((((............)))).).))))))..(((((((((((........))))))))))) (-20.41 = -20.13 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:53 2006