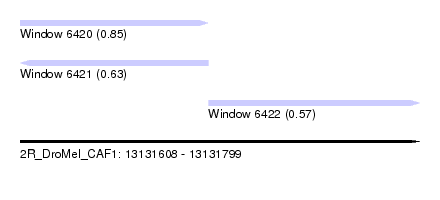
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,131,608 – 13,131,799 |
| Length | 191 |
| Max. P | 0.854332 |

| Location | 13,131,608 – 13,131,698 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -23.08 |
| Consensus MFE | -18.48 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
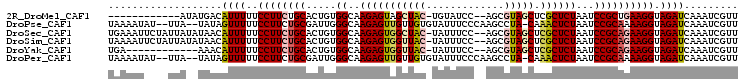
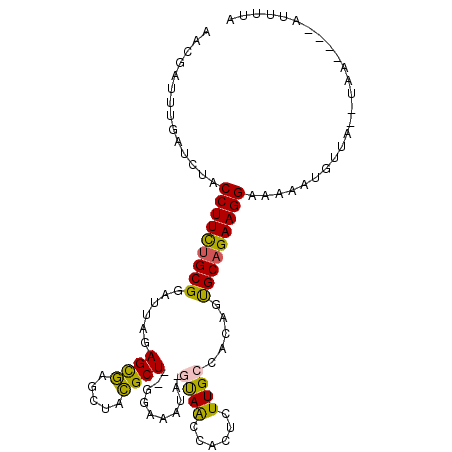
>2R_DroMel_CAF1 13131608 90 + 20766785 ------------AUAUGACAUUUUUCCUUCUGCACUGUGGCAAGAGUAGCUAC-UGUAUCC--AGCGUAGCUCGCUCUAAUCCGCUGAAGGUAGAUCAAAUCGUU ------------..((((....((((((((......((((..(((((((((((-.((....--.)))))))).)))))...)))).))))).))).....)))). ( -24.90) >DroPse_CAF1 25453 100 + 1 UAAAAUAU--UUA--UAUAGUUUUUCCUUCUGCGAUUGGGCAAGAGUUGUUGUGUAUUUCCCAAGCCUA-CAAACUCUAAUCCGCAAAAGGUAGAUCAAAUCGUU ........--...--....(.(((.((((.(((....(((..((((((((.(.((.........))).)-).))))))..)))))).)))).))).)........ ( -15.70) >DroSec_CAF1 25474 102 + 1 UGAAAUUCUAUUAUAUAACAUUUUUCCUUCUGCACUGUGGCAAGAGUGGCUAC-UAUUUCC--AGCGUAGCUCGCUCUAAUCCGCAGAAGGUAGAUCAAAUCGUU ............................(((((.((((((..(((((((((((-.......--...))))).))))))...))))))...))))).......... ( -29.00) >DroSim_CAF1 27362 102 + 1 UAAAAUUCUAUUAUAUAACAUUUUUCCUUCUGCACUGUGGCAAGAGUGGUUAC-UAUUUCC--AGCGUAGCUCGCUCUAAUCCGCAGAAGGUAGAUCAAAUCGUU ............................(((((.((((((..(((((((((((-.......--...))))).))))))...))))))...))))).......... ( -26.60) >DroYak_CAF1 25821 90 + 1 UGA------------AAACAUUUUUCCUUCUGCACUGUGGCAAGAGUGGUUAC-UAUUUCC--AGCGUAGCUCGCUCUAAUCCGCAGAAGGUAGAUCAAAUCGUU ...------------.............(((((.((((((..(((((((((((-.......--...))))).))))))...))))))...))))).......... ( -26.60) >DroPer_CAF1 25393 100 + 1 UAAAAUAU--UUA--UAUAGUUUUUCCUUCUGCGAUUGGGCAAGAGUUGUUGUGUAUUUCCCAAGCCUA-CAAACUCUAAUCCGCAAAAGGUAGAUCAAAUCGUU ........--...--....(.(((.((((.(((....(((..((((((((.(.((.........))).)-).))))))..)))))).)))).))).)........ ( -15.70) >consensus UAAAAU____UUA__UAACAUUUUUCCUUCUGCACUGUGGCAAGAGUGGUUAC_UAUUUCC__AGCGUAGCUCGCUCUAAUCCGCAGAAGGUAGAUCAAAUCGUU ...................((((..((((((((.....((..(((((((((((.............))))).))))))...)))))))))).))))......... (-18.48 = -18.70 + 0.23)
| Location | 13,131,608 – 13,131,698 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -22.09 |
| Consensus MFE | -16.55 |
| Energy contribution | -15.50 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13131608 90 - 20766785 AACGAUUUGAUCUACCUUCAGCGGAUUAGAGCGAGCUACGCU--GGAUACA-GUAGCUACUCUUGCCACAGUGCAGAAGGAAAAAUGUCAUAU------------ ...(((........(((((.((((...((((..((((..(((--(....))-)))))).))))..)).....)).)))))......)))....------------ ( -22.54) >DroPse_CAF1 25453 100 - 1 AACGAUUUGAUCUACCUUUUGCGGAUUAGAGUUUG-UAGGCUUGGGAAAUACACAACAACUCUUGCCCAAUCGCAGAAGGAAAAACUAUA--UAA--AUAUUUUA ..............((((((((((...((((((((-((..(....)...))))....)))))).......))))))))))..........--...--........ ( -20.10) >DroSec_CAF1 25474 102 - 1 AACGAUUUGAUCUACCUUCUGCGGAUUAGAGCGAGCUACGCU--GGAAAUA-GUAGCCACUCUUGCCACAGUGCAGAAGGAAAAAUGUUAUAUAAUAGAAUUUCA ..............((((((((((...((((.(.(((((...--.......-)))))).))))..)).....))))))))......................... ( -27.20) >DroSim_CAF1 27362 102 - 1 AACGAUUUGAUCUACCUUCUGCGGAUUAGAGCGAGCUACGCU--GGAAAUA-GUAACCACUCUUGCCACAGUGCAGAAGGAAAAAUGUUAUAUAAUAGAAUUUUA ..............(((((((((......((((.....))))--((.((.(-((....))).)).))....)))))))))......................... ( -21.30) >DroYak_CAF1 25821 90 - 1 AACGAUUUGAUCUACCUUCUGCGGAUUAGAGCGAGCUACGCU--GGAAAUA-GUAACCACUCUUGCCACAGUGCAGAAGGAAAAAUGUUU------------UCA ..............(((((((((......((((.....))))--((.((.(-((....))).)).))....)))))))))..........------------... ( -21.30) >DroPer_CAF1 25393 100 - 1 AACGAUUUGAUCUACCUUUUGCGGAUUAGAGUUUG-UAGGCUUGGGAAAUACACAACAACUCUUGCCCAAUCGCAGAAGGAAAAACUAUA--UAA--AUAUUUUA ..............((((((((((...((((((((-((..(....)...))))....)))))).......))))))))))..........--...--........ ( -20.10) >consensus AACGAUUUGAUCUACCUUCUGCGGAUUAGAGCGAGCUACGCU__GGAAAUA_GUAACCACUCUUGCCACAGUGCAGAAGGAAAAAUGUUA__UAA____AUUUUA ..............(((((((((......((((.....))))..........((((......)))).....)))))))))......................... (-16.55 = -15.50 + -1.05)

| Location | 13,131,698 – 13,131,799 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 78.46 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -16.20 |
| Energy contribution | -15.95 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
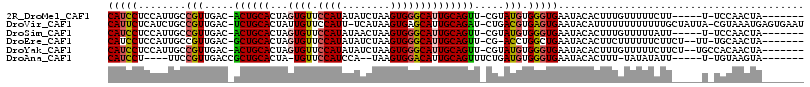
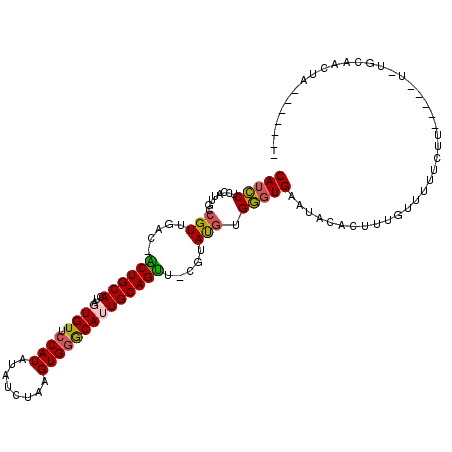
>2R_DroMel_CAF1 13131698 101 + 20766785 CAUCCUCCAUUGCCGUUGAC-ACUGCACUAGUGUUCCAUAUAUCUAAGUGGGCAUUGCAGUU-CGUAUGUGGGUGAAUACACUUUGUUUUUCUU-----U-UCCAACUA------- ..((.(((((......(((.-((((((...((((.((((........)))))))))))))))-))...))))).))..................-----.-........------- ( -20.30) >DroVir_CAF1 31297 112 + 1 CAUUCUCAUCUGCCGUUGAC-UCUGCACUAUUGUUCCAUU-UCAUAAGUGAGCAUUGCAGAU-CUGACGUGAGUGAAUACAUUUUUUUUUUUUGCUAUUA-CGUAAAUGAGUGAAU ((((((((.((..(((((..-((((((....(((((.(((-.....)))))))).)))))).-.)))))..)))))..............(((((.....-.))))).)))))... ( -24.40) >DroSim_CAF1 27464 101 + 1 CAUCCUCCAUUGCCGUUGAC-ACUGCACUAGUGUUCCAUAUAACUAAGUGGGCAUUGCAGUU-CGUAUGUGGGUGAAUACACUUUGUUUUUAUU-----U-UCCAACUA------- ..((.(((((......(((.-((((((...((((.((((........)))))))))))))))-))...))))).))..................-----.-........------- ( -20.30) >DroEre_CAF1 25894 103 + 1 CAUCCUCCAUUGCCGUUGAC-GCUGCACUAGUGUUCCAUAUAUCUAAGUGGGCAUUGCAGUU-CG-ACCUGGCUGAAUACACUUCUUUUUUCUUCU--UU-UGCAACUA------- ...........((((((((.-((((((...((((.((((........)))))))))))))))-))-))..))).......................--..-........------- ( -22.60) >DroYak_CAF1 25911 105 + 1 CAUCCUCCAUUGCCGUUGAC-ACUGCACUAGUGUUCCAUAUAUCUAAGUGGGCAUUGCAGUU-CGUAUGUGGGUGAAUACACUUUGUUUUUCUUCU--UGCCACAACUA------- ..............((((..-((((((...((((.((((........)))))))))))))).-.....(..((.(((.((.....))..)))..))--..)..))))..------- ( -21.30) >DroAna_CAF1 25864 95 + 1 CAUCCU----UUCCGUUGACCGCUGCACUA-UGUUCCAUCCA--UAAGUGGACAUUGCAGUUUCUGAUGUGGGUGAAUACACUUU-UAUAUAUU-----U-UGUAAGUA------- (((((.----...(((((...((((((..(-(((.((((...--...))))))))))))))...))))).)))))........((-((((....-----.-))))))..------- ( -22.20) >consensus CAUCCUCCAUUGCCGUUGAC_ACUGCACUAGUGUUCCAUAUAUCUAAGUGGGCAUUGCAGUU_CGUAUGUGGGUGAAUACACUUUGUUUUUCUU_____U_UGCAACUA_______ (((((........(((.....((((((...((((.((((........)))))))))))))).....))).)))))......................................... (-16.20 = -15.95 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:49 2006