| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,127,595 – 13,127,690 |
| Length | 95 |
| Max. P | 0.997324 |

| Location | 13,127,595 – 13,127,690 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 73.42 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.54 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997324 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
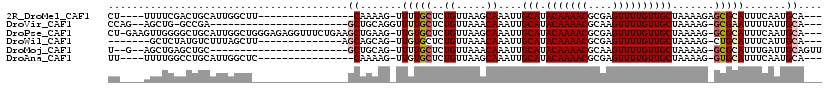
>2R_DroMel_CAF1 13127595 95 + 20766785 CU----UUUUCGACUGCAUUGGCUU----------------CAAAAG-UUUUGCUCUGUUAAGCAAAUUGCAUACAAAACGCGAGUUUUGUUGCUAAAAGAGCGCAUUUCAAUGCA--- ..----.........(((((((...----------------......-.((((((......)))))).(((..(((((((....))))))).(((.....))))))..))))))).--- ( -26.40) >DroVir_CAF1 26758 89 + 1 CCAG--AGCUG-GCCGA-----------------------GCUGCAGGUUUUGCUCUGUUAAACAAAUUGCAUACAAAACGCAAGUUUUGUUGCUAAAAG-GCGAAUUUUAUUGCA--- .(((--(((.(-(((..-----------------------......))))..))))))...........(((.(((((((....))))))))))......-((((......)))).--- ( -29.20) >DroPse_CAF1 21650 113 + 1 CU-GAAGUUGGGGCUGCAUUGGCUGGGAGAGGUUUCUGAAGCUGAAG-UUGUGCUCUGUUAAGCAAAUUGCAUACAAAACGCGAGUUUUGUUGCUAAAAG-GCGCAUUUCAAUGCA--- ..-......(((((.((.(..(((.((((....))))..)))..).)-)...))))).....((.....(((.(((((((....))))))))))......-))((((....)))).--- ( -33.60) >DroWil_CAF1 23141 93 + 1 -------GCUCUAUGUCUUUAGCUU--------------AGCAGCAG-UUGUGCUCUGUUAAACAAAUUGCAUACAAAACGCGAGUUUUGUUGCUAAAAG-CUGCAUUUCAUUGCA--- -------((...(((....((((((--------------((((((((-((((..........)).))))))..(((((((....))))))))))))..))-))).....))).)).--- ( -26.80) >DroMoj_CAF1 24117 90 + 1 U--G--AGCUGAGCUGC-----------------------GCUGCAG-UUUUGCUCUGUUAAACAAAUUGCAUACAAAACGCAAGUUUUGUUGCUAAAAG-GCGCAUUUGAUUGCAGUU .--(--(((.(((((((-----------------------...))))-))).))))(((....(((((.((..(((((((....))))))).(((....)-)))))))))...)))... ( -31.40) >DroAna_CAF1 21678 94 + 1 UU----UUUUGGCCUGCAUUGGCUC----------------CAAAAG-UUGUGCUCUGUUAAGCAAAUUGCAUACAAAACGCGAGUUUUGUUGCUAAAAG-GUGCAUUUCAAUGCA--- ..----.........((((((((.(----------------(.....-...((((......))))....(((.(((((((....)))))))))).....)-).))....)))))).--- ( -27.00) >consensus CU____AGCUGGGCUGCAUUGGCU________________GCAGAAG_UUGUGCUCUGUUAAACAAAUUGCAUACAAAACGCGAGUUUUGUUGCUAAAAG_GCGCAUUUCAAUGCA___ ........................................((.......(((((..((.....))....(((.(((((((....)))))))))).......))))).......)).... (-14.10 = -14.54 + 0.44)

| Location | 13,127,595 – 13,127,690 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.42 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -13.94 |
| Energy contribution | -13.63 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
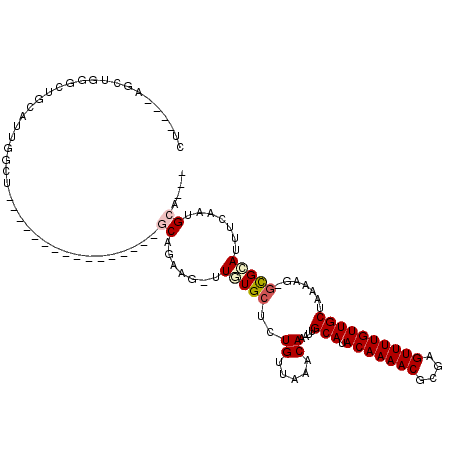
>2R_DroMel_CAF1 13127595 95 - 20766785 ---UGCAUUGAAAUGCGCUCUUUUAGCAACAAAACUCGCGUUUUGUAUGCAAUUUGCUUAACAGAGCAAAA-CUUUUG----------------AAGCCAAUGCAGUCGAAAA----AG ---(((((((...((((((.....))).(((((((....)))))))..))).(((((((....))))))).-......----------------....)))))))........----.. ( -25.60) >DroVir_CAF1 26758 89 - 1 ---UGCAAUAAAAUUCGC-CUUUUAGCAACAAAACUUGCGUUUUGUAUGCAAUUUGUUUAACAGAGCAAAACCUGCAGC-----------------------UCGGC-CAGCU--CUGG ---((((.........((-......)).(((((((....))))))).))))..........((((((....((......-----------------------..)).-..)))--))). ( -20.70) >DroPse_CAF1 21650 113 - 1 ---UGCAUUGAAAUGCGC-CUUUUAGCAACAAAACUCGCGUUUUGUAUGCAAUUUGCUUAACAGAGCACAA-CUUCAGCUUCAGAAACCUCUCCCAGCCAAUGCAGCCCCAACUUC-AG ---(((((((...(((((-......)).(((((((....)))))))..)))...(((((....)))))...-.....(((..(((....)))...))))))))))...........-.. ( -24.30) >DroWil_CAF1 23141 93 - 1 ---UGCAAUGAAAUGCAG-CUUUUAGCAACAAAACUCGCGUUUUGUAUGCAAUUUGUUUAACAGAGCACAA-CUGCUGCU--------------AAGCUAAAGACAUAGAGC------- ---.(((......)))((-(((.((((((((((((....))))))).(((..((((.....)))))))...-.)))))..--------------))))).............------- ( -21.00) >DroMoj_CAF1 24117 90 - 1 AACUGCAAUCAAAUGCGC-CUUUUAGCAACAAAACUUGCGUUUUGUAUGCAAUUUGUUUAACAGAGCAAAA-CUGCAGC-----------------------GCAGCUCAGCU--C--A ....((.......(((((-......((((((((((....))))))).)))..(((((((....))))))).-.....))-----------------------))).....)).--.--. ( -22.30) >DroAna_CAF1 21678 94 - 1 ---UGCAUUGAAAUGCAC-CUUUUAGCAACAAAACUCGCGUUUUGUAUGCAAUUUGCUUAACAGAGCACAA-CUUUUG----------------GAGCCAAUGCAGGCCAAAA----AA ---(((((((....((.(-(.....((((((((((....))))))).)))....(((((....)))))...-.....)----------------).)))))))))........----.. ( -24.80) >consensus ___UGCAAUGAAAUGCGC_CUUUUAGCAACAAAACUCGCGUUUUGUAUGCAAUUUGCUUAACAGAGCAAAA_CUGCAGC________________AGCCAAUGCAGCCCAAAA____AG ...((((......))))........((((((((((....))))))).)))....(((((....)))))................................................... (-13.94 = -13.63 + -0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:46 2006