| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,378,484 – 2,378,593 |
| Length | 109 |
| Max. P | 0.500000 |

| Location | 2,378,484 – 2,378,593 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -23.97 |
| Energy contribution | -23.58 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
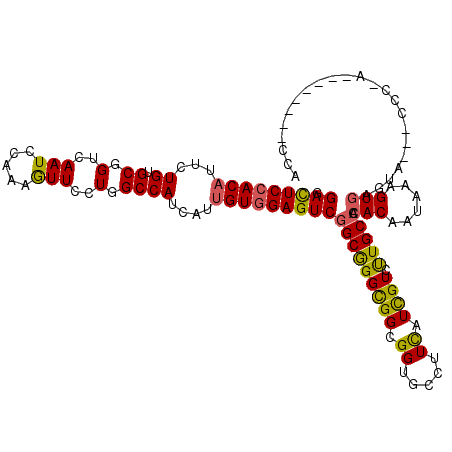
>2R_DroMel_CAF1 2378484 109 - 20766785 GGACUCCACAUUCUGCGCGGUGAAUCCCAAGUUCCUGGCCAUUAUCGUAGAGUCGGCGGGCGGCGGAGCCUUCAUCGUCCUGCCACACAACAAAGUGAGUG---CGCCA--------CCG .(((((.((((..((.((.(.((((.....)))).).))))..)).)).)))))((((.(((((((..(.......)..))))).(((......))).)).---)))).--------... ( -37.00) >DroVir_CAF1 7195 107 - 1 GGAUUCCACAUUCUGUGCGGUCAAUCCAAAGUUUCUGGCCAUCAUUGUGGAGUCGGCGGGCGGCGGCGCGUUCAUUGUAUUGCCACACAAUAAAGUGAGUA---CCG----------GCA .(((((((((...((...(((((............)))))..)).)))))))))(.((((((((((..((.....))..))))).(((......))).)).---)))----------.). ( -36.60) >DroGri_CAF1 4969 117 - 1 GGACUCCACAUUCUGUGCGGUCAAUCCAAAAUUCUUGGCCAUAAUUGUGGAGUCGGCGGGCGGCGGUGCAUUCAUUGUGUUGCCACACAAUAAAGUGAGUA---UUAUAAUUAUUAAUCA .(((((((((.....((.((((((..........)))))).))..))))))))).....(((((((..((.....))..))))).(((......))).)).---................ ( -35.30) >DroMoj_CAF1 6630 110 - 1 GGACUCCACAUUCUGUGCGGUAAAUCCAAAGUUUUUGGCCAUCAUUGUGGAGUCAGCAGGUGGCGGAGCCUUCAUCGUAUUGCCACACAAUAAAGUGAGUAUAUCUG----------AUA .(((((((((...((.((.(.((((.....)))).).))))....)))))))))..((((((((...)))(((((..(((((.....)))))..)))))...)))))----------... ( -32.10) >DroAna_CAF1 3820 109 - 1 GGACUCCACAUUCUGUGCCGUCAAUCCAAAGUUCCUGGCCAUCAUUGUGGAGUCGGCGGGCGGCGGUGCCUUCAUCGUCUUGCCCCACAACAAAGUAAGUG---CCCCA--------UUC .(((((((((...((.((((..(((.....)))..))))))....)))))))))((((((((((((((....)))))))..)))).((......))....)---))...--------... ( -39.50) >DroPer_CAF1 3874 111 - 1 GGACUCCACAUUCUGCGCCGUCAAUCCAAAGUUCCUGGCCAUCAUUGUGGAGUCGGCGGGCGGUGGUGCCUUUAUCGUCUUGCCACACAAUAAAGUGAGUAUG-CACAG--------CCA .(((((((((...((.((((..(((.....)))..))))))....)))))))))((((((((((((.....)))))))))((((((........))).)))..-....)--------)). ( -39.00) >consensus GGACUCCACAUUCUGUGCGGUCAAUCCAAAGUUCCUGGCCAUCAUUGUGGAGUCGGCGGGCGGCGGUGCCUUCAUCGUCUUGCCACACAAUAAAGUGAGUA___CCC_A________CCA .(((((((((...((.((.(..(((.....)))..).))))....)))))))))(((((((((.((.....)).)))).))))).(((......)))....................... (-23.97 = -23.58 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:31 2006