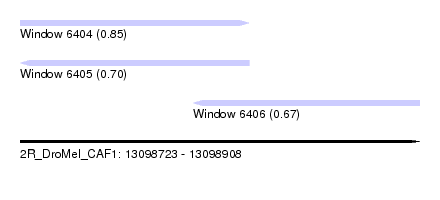
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,098,723 – 13,098,908 |
| Length | 185 |
| Max. P | 0.846000 |

| Location | 13,098,723 – 13,098,829 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.82 |
| Mean single sequence MFE | -36.33 |
| Consensus MFE | -32.39 |
| Energy contribution | -32.73 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13098723 106 + 20766785 CCAAUGGAGGCGCCCAUGCAGAGGGCACCAAAGCUGGCCGACAAGCCAGCAAUAUAUUGUGGCAACACCGAUCCCUUGGACAUGGCGAGGGGUUUUUUUUUUAAAU-------------- ........((.((((.......)))).))...((((((......))))))........(((....))).(((((((((.......)))))))))............-------------- ( -38.40) >DroSim_CAF1 9899 120 + 1 CCGAUGGAGGCGCCCAUGCAGAGGGCACCAAAACUGGCCGACAAGCCAGCAAUAUAUUGUGGCAACACCGAUCCCUUGGACAUGGCGAGGGAUUUUUUUUUUGAAUUUGAAUUUUUUGAU ........((.((((.......)))).))....(((((......)))))(((...((((((....))).(((((((((.......)))))))))...............)))...))).. ( -34.90) >DroYak_CAF1 10219 103 + 1 CCGAUGGAGGCCCCCAUGCAGAGGGCACCAAAGCUGGCCGACAAGCCGGCAAUAUAUUGUGGCAAUACCGAUCCCUUGGACAUGGCGAGGGAUGUAU---UAGAAA-------------- (((.(((..((((.(.....).)))).)))..((((((......)))))).........))).(((((..((((((((.......))))))))))))---).....-------------- ( -35.70) >consensus CCGAUGGAGGCGCCCAUGCAGAGGGCACCAAAGCUGGCCGACAAGCCAGCAAUAUAUUGUGGCAACACCGAUCCCUUGGACAUGGCGAGGGAUUUUUUUUUUGAAU______________ ........((.((((.......)))).))...((((((......))))))........(((....))).(((((((((.......))))))))).......................... (-32.39 = -32.73 + 0.34)


| Location | 13,098,723 – 13,098,829 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.82 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -29.84 |
| Energy contribution | -30.73 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13098723 106 - 20766785 --------------AUUUAAAAAAAAAACCCCUCGCCAUGUCCAAGGGAUCGGUGUUGCCACAAUAUAUUGCUGGCUUGUCGGCCAGCUUUGGUGCCCUCUGCAUGGGCGCCUCCAUUGG --------------...................((((..((((...)))).))))......((((.....(((((((....)))))))...(((((((.......)))))))...)))). ( -34.70) >DroSim_CAF1 9899 120 - 1 AUCAAAAAAUUCAAAUUCAAAAAAAAAAUCCCUCGCCAUGUCCAAGGGAUCGGUGUUGCCACAAUAUAUUGCUGGCUUGUCGGCCAGUUUUGGUGCCCUCUGCAUGGGCGCCUCCAUCGG ...........................((((((.(.......).))))))(((((...............(((((((....)))))))...(((((((.......)))))))..))))). ( -35.20) >DroYak_CAF1 10219 103 - 1 --------------UUUCUA---AUACAUCCCUCGCCAUGUCCAAGGGAUCGGUAUUGCCACAAUAUAUUGCCGGCUUGUCGGCCAGCUUUGGUGCCCUCUGCAUGGGGGCCUCCAUCGG --------------....((---((((((((((.(.......).))))))..))))))((..........((.((((....)))).))..(((.((((((.....))))))..)))..)) ( -32.10) >consensus ______________AUUCAAAAAAAAAAUCCCUCGCCAUGUCCAAGGGAUCGGUGUUGCCACAAUAUAUUGCUGGCUUGUCGGCCAGCUUUGGUGCCCUCUGCAUGGGCGCCUCCAUCGG ...........................((((((.(.......).))))))(((((...............(((((((....)))))))...(((((((.......)))))))..))))). (-29.84 = -30.73 + 0.89)

| Location | 13,098,803 – 13,098,908 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -20.53 |
| Energy contribution | -20.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13098803 105 - 20766785 AUUUUUCCAGCGUAAUAACGGUCGCGUGGCGAAUUCAGUUCGUUUCCGGCGAUCGACAAGGAAACACCUAUAAAAACCA---------------AUUUAAAAAAAAAACCCCUCGCCAUG .........(((......(((((((..(((((((...)))))))....)))))))....(....)..............---------------...................))).... ( -20.60) >DroSim_CAF1 9979 120 - 1 AUUUUUCCAGCGUAAUAACGGUCGCGUGGCGAAUUCAGUUCGUUUCCGGCGAUCGAGAGGGAAACACCUAUAAAAACCAUAUCAAAAAAUUCAAAUUCAAAAAAAAAAUCCCUCGCCAUG .........((.......(((((((..(((((((...)))))))....))))))).((((((..............................................)))))))).... ( -25.35) >DroYak_CAF1 10299 102 - 1 AUUUUUCCAGCGUAAUAACGGUCGCGUGGCGAAUUCAGUUCGUUUCCGGCGAUCGAGAAGGAAACACCUAUACAAAAUA---------------UUUCUA---AUACAUCCCUCGCCAUG .........(((......(((((((..(((((((...)))))))....)))))))....(....)..............---------------......---..........))).... ( -20.60) >consensus AUUUUUCCAGCGUAAUAACGGUCGCGUGGCGAAUUCAGUUCGUUUCCGGCGAUCGAGAAGGAAACACCUAUAAAAACCA_______________AUUCAAAAAAAAAAUCCCUCGCCAUG .........(((......(((((((..(((((((...)))))))....)))))))....(....)................................................))).... (-20.53 = -20.53 + 0.00)

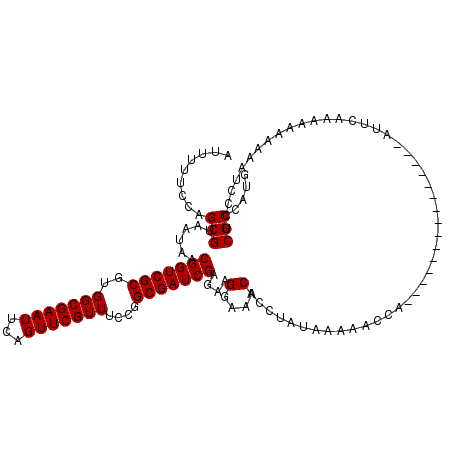
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:33 2006