| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,054,642 – 13,054,852 |
| Length | 210 |
| Max. P | 0.513050 |

| Location | 13,054,642 – 13,054,738 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 79.86 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -17.47 |
| Energy contribution | -17.20 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
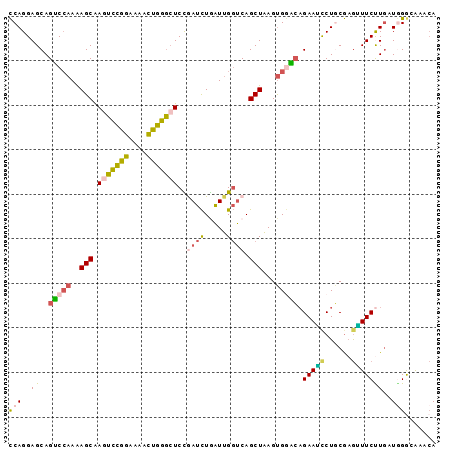
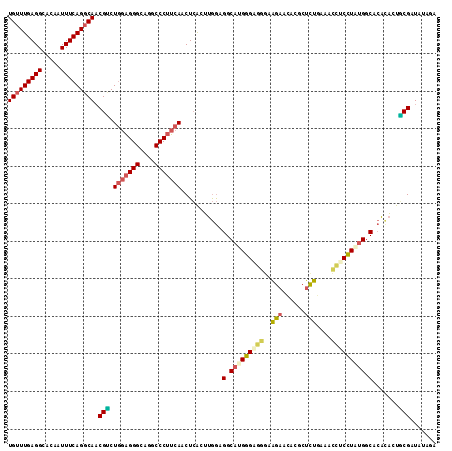
>2R_DroMel_CAF1 13054642 96 + 20766785 CCAGGAGCAGUCCAAAAGCAAGUCCGGAAAAUUGGGCUCUGAUCUGAUUGGUCAGCUAAGUGGACAGAAGCCUGCGAGUUUCUUGAUGGGGAAACA .((((..(.(((((..(((.(((((((....))))))).(((((.....))))))))...))))).)...))))...((((((......)))))). ( -32.40) >DroSec_CAF1 3390 96 + 1 CCAGGAGCAGUCCAAAAGCAAGUCCGGUAAAUUGGGAUCCGAUCUGAUUGGUCAGCUAAGUGGACAGAAACCUGCGAGUUUCUUGAUGGGGAAACA .((((..(.(((((..(((..((((.(.....).))))..((((.....)))).)))...))))).)...))))...((((((......)))))). ( -28.90) >DroSim_CAF1 3342 96 + 1 CCAGGAGCAGUCCAAAAGCAAGUCCGGUAAAUUGGGCUCCGAUCUGAUUGGUCAGCUAAGUGGACAGAAGCCUGCGAGUUUCUUGAUGGGUAAACA (((....(((((((..(((.(((((((....)))))))..((((.....)))).)))...)))))((((((......)))))))).)))....... ( -28.60) >DroEre_CAF1 3332 96 + 1 CCAGGAGCAGUCGAAAAGCGAGUCCGGAAAACUGGGCUCUAAUCUAAUUGGUCAGCUAAGUGGGCAGAAUCCUGCGAGAUUCCUGAUGGACAAACA (((..((.((((.......((((((((....))))))))....(((.((((....)))).)))((((....))))..)))).))..)))....... ( -28.80) >DroYak_CAF1 3314 96 + 1 CCAGGAGCAGUCCAAAAGCGAGUCCGGAAAACUGGGCUCCAAUCUGAUUGGUCAGCUAAGUGCGCAGAAUUCUACGAGAUUCUUAAUAGGAAAACA ((..((.(((((.....(.((((((((....))))))))).....))))).)).((.....))..((((((......)))))).....))...... ( -24.90) >DroAna_CAF1 3220 96 + 1 UCAGGAGCAGGCCAAGAGCGACUUUAGUAAACUGAACUCUGACUUAGUGGCCCAGCUCAGCGAUGUGAAUCCUCCACAGUUCCUGAUAGACGGGGA ((((((((.(((((.((((((.(((((....))))).)).).)))..)))))...........((((.......)))))))))))).......... ( -29.30) >consensus CCAGGAGCAGUCCAAAAGCAAGUCCGGAAAACUGGGCUCCGAUCUGAUUGGUCAGCUAAGUGGACAGAAUCCUGCGAGUUUCUUGAUGGGCAAACA (((..((..(((((..(((.(((((((....)))))))..((((.....)))).)))...))))).(((((......)))))))..)))....... (-17.47 = -17.20 + -0.27)


| Location | 13,054,738 – 13,054,852 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -39.57 |
| Consensus MFE | -29.51 |
| Energy contribution | -29.98 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
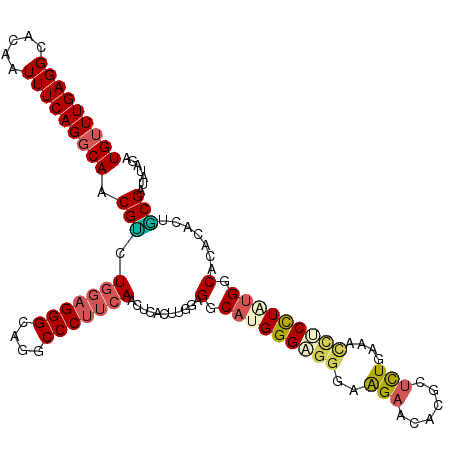
>2R_DroMel_CAF1 13054738 114 - 20766785 UGUUUGAGGCACAAUUUCAGGCAACGUCUGGAGGGCAGGCCCUUCAACUCACUUGGAGGCAUGGGAGGGAAGAACACGCUCUGAAAUCUCCUAUGGCACACACUGCGAUAUAGA (((((((((.....)))))))))..((((((((((....))))))).(((.....))).(((((((((..(((......)))....)))))))))(((.....))))))..... ( -35.70) >DroVir_CAF1 5109 114 - 1 UGCUUGAGGCACAGUUUCAGACAACGACUCGUGGGCAGUCCCUGAAACUGAGCGAACGGCACGGGCGCCAGGAAGACGCCUUUAAACAGCCUCGGACAAUUGCUUCGAUACAGA ...(((((((((((((((((...((..((....))..))..)))))))))..(((..(((..(((((.(.....).))))).......))))))......))))))))...... ( -34.10) >DroSec_CAF1 3486 114 - 1 UGUUUGAGGCACAAUUUCAGGCAACGUCUGGAGGGCAGGCCCUUCAACUCACUUGGUGGCAUGGGAGGGAAGAACACGCUCUGAAACCUCCUAUGGCACACAGUACGAUAUAGA (((((((((.....))))))))).(((.(((((((....)))))))....(((..(((.(((((((((..(((......)))....))))))))).)))..))))))....... ( -43.00) >DroSim_CAF1 3438 114 - 1 UGUUUGAGGCAUAAUUUCAGGCAACGUCUGGAGGGCAGGCCCUUCAACUCACUUGGUGGCAUGGGAGGGAAGAACACGCUCUGAAACCUCCUAUGGCACACAGUGCGAUAUAGA (((((((((.....)))))))))..((((((((((....)))))))...((((..(((.(((((((((..(((......)))....))))))))).)))..)))).)))..... ( -44.90) >DroEre_CAF1 3428 114 - 1 UGUUUGAGGCACAAUUUCAGGCAUCGCCUGGAGGGCAGACCCUUCAACUCGCUUGGAGGCAGGGGUGGGAAGAACACGCUUUGAAACUUCCUAUGGCACACACUGCGAUAUAGA .((((((((.....))))))))(((((.(((((((....)))))))....(((.(((((((((((((.......))).)))))...)))))...))).......)))))..... ( -37.20) >DroYak_CAF1 3410 114 - 1 UGUUUGAGGCACAAUUUCAGGCAACGCCUGGAGGGCAGGCCCUUCAACUCACUUGGCGGCAUGGGUGGGAAGAAGACGCUCUGAAACCUCUUGUGGCACAAACUGCGAUAAAGA .(((((.(.(((((...(((((...)))))((((.(((((.((((..((((((((......))))))))..))))..)).)))...))))))))).).)))))........... ( -42.50) >consensus UGUUUGAGGCACAAUUUCAGGCAACGUCUGGAGGGCAGGCCCUUCAACUCACUUGGAGGCAUGGGAGGGAAGAACACGCUCUGAAACCUCCUAUGGCACACACUGCGAUAUAGA (((((((((.....))))))))).(((.(((((((....)))))))...........(.(((((((((..(((......)))....))))))))).).......)))....... (-29.51 = -29.98 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:19 2006