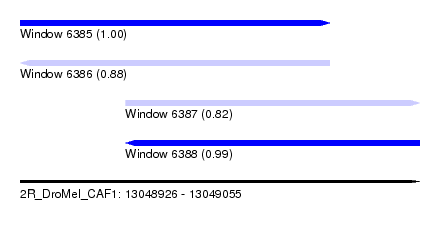
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,048,926 – 13,049,055 |
| Length | 129 |
| Max. P | 0.996237 |

| Location | 13,048,926 – 13,049,026 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -26.20 |
| Energy contribution | -26.84 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13048926 100 + 20766785 GAGAAUGGGCAGCGAUAUCUGCGGCUAAAUAUAACAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGAGAGUGCUCCGCUUGAGUCAGCGCUUGGAUGU-------------- .......(((.(((.....))).))).......(((..((((((((((......(((((((((.........))).))))))..))))))))))...)))-------------- ( -33.10) >DroSec_CAF1 19348 100 + 1 GAGAAUGGGCAGCGAUAUCUGCGGCUAAAUAUAACAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGAGAGUGCUCCCCUUGAGUCAGCGCCUCGAUGU-------------- (((....(((.(((.....))).)))..............((((((((.((((..((((..(.......)..))))..))))..))))))))))).....-------------- ( -29.80) >DroSim_CAF1 19063 100 + 1 GAGAAUGGGCAGCGAUAUCUUCGGCUAAAUAUAACAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGAGAGUGCCCCGCUUGAGUCAGCGCCUCGAUGU-------------- (((....(((..(((.....))))))..............((((((((......(((((((((.........)).)))))))..))))))))))).....-------------- ( -30.40) >DroEre_CAF1 21061 100 + 1 GAGAAUGGGAAGCGAUAUCUGCGGCUAAAUAUAACAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGAGAGUGCUCCGCUUGAGUCAGCGACUCGAUGU-------------- .......((..(((.....)))..)).......(((..((((((((((......(((((((((.........))).))))))..)))))).))))..)))-------------- ( -29.90) >DroYak_CAF1 21693 114 + 1 GAGAAUGGGCAGCGAUAUCUGCGGCUAAAUAUAACAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGUGAGUGCUCCGCUUAAGUCAGCGACUCGAUGUUUAAAUGUUUGAAU .......(((.(((.....))).)))((((((((((..((((((((((......(((((((((.........))).))))))..)))))).))))..))))...)))))).... ( -33.90) >consensus GAGAAUGGGCAGCGAUAUCUGCGGCUAAAUAUAACAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGAGAGUGCUCCGCUUGAGUCAGCGCCUCGAUGU______________ .......(((.(((.....))).))).......(((..((((((((((......(((((((((.........))).))))))..))))))).)))..))).............. (-26.20 = -26.84 + 0.64)

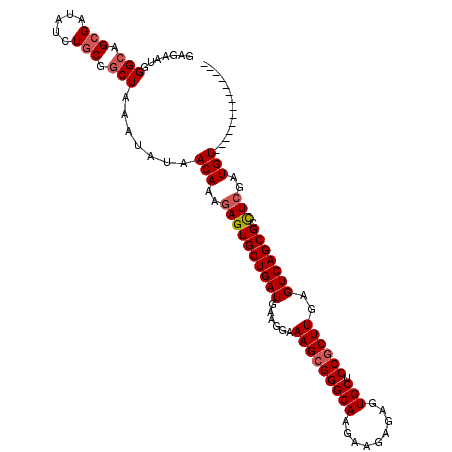
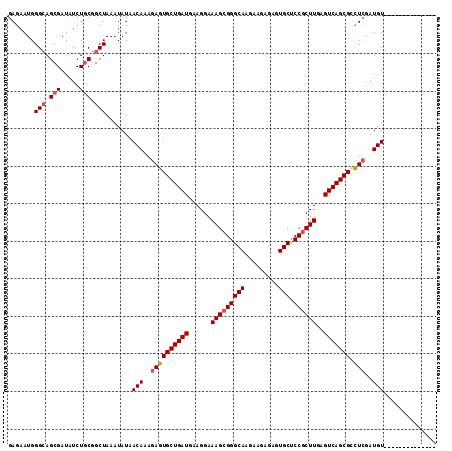
| Location | 13,048,926 – 13,049,026 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -26.15 |
| Consensus MFE | -18.52 |
| Energy contribution | -19.52 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13048926 100 - 20766785 --------------ACAUCCAAGCGCUGACUCAAGCGGAGCACUCUCUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUGUUAUAUUUAGCCGCAGAUAUCGCUGCCCAUUCUC --------------.......((.(((((...((((((.(((.........))))))))).......))))).))....((((....)))).((((......))))........ ( -22.70) >DroSec_CAF1 19348 100 - 1 --------------ACAUCGAGGCGCUGACUCAAGGGGAGCACUCUCUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUGUUAUAUUUAGCCGCAGAUAUCGCUGCCCAUUCUC --------------(((..(((..(((((...((((..(((...............)))..))))..))))).)))..)))...........((((......))))........ ( -27.56) >DroSim_CAF1 19063 100 - 1 --------------ACAUCGAGGCGCUGACUCAAGCGGGGCACUCUCUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUGUUAUAUUUAGCCGAAGAUAUCGCUGCCCAUUCUC --------------.....(.((((((((...(((((((.((.........))))))))).......)))))..(((((((((....))).)))))).......))))...... ( -26.70) >DroEre_CAF1 21061 100 - 1 --------------ACAUCGAGUCGCUGACUCAAGCGGAGCACUCUCUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUGUUAUAUUUAGCCGCAGAUAUCGCUUCCCAUUCUC --------------.....((((.(((((...((((((.(((.........))))))))).......)))))))))...((.((((((......)))))).))........... ( -23.80) >DroYak_CAF1 21693 114 - 1 AUUCAAACAUUUAAACAUCGAGUCGCUGACUUAAGCGGAGCACUCACUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUGUUAUAUUUAGCCGCAGAUAUCGCUGCCCAUUCUC .............((((..((((.(((((...((((((.(((.........))))))))).......)))))))))..))))..........((((......))))........ ( -30.00) >consensus ______________ACAUCGAGGCGCUGACUCAAGCGGAGCACUCUCUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUGUUAUAUUUAGCCGCAGAUAUCGCUGCCCAUUCUC ..............(((..(((..(((((...((((((.(((.........))))))))).......))))).)))..)))...........((((......))))........ (-18.52 = -19.52 + 1.00)

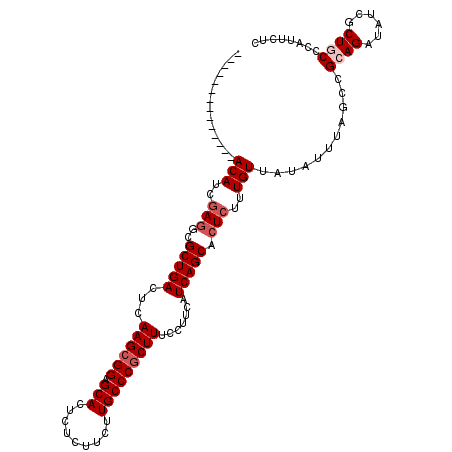
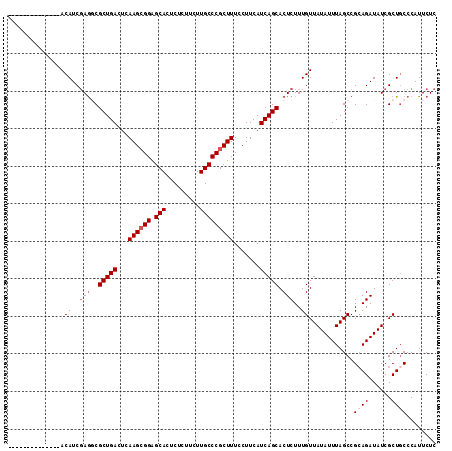
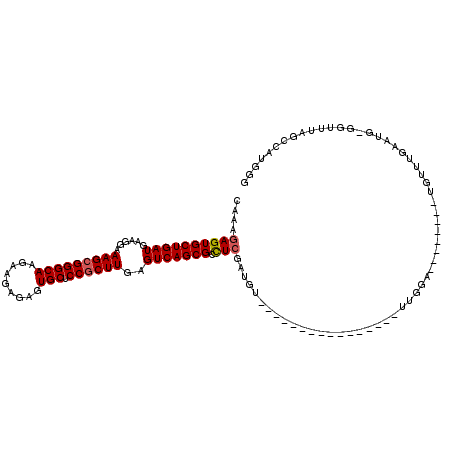
| Location | 13,048,960 – 13,049,055 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.33 |
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -21.76 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13048960 95 + 20766785 CAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGAGAGUGCUCCGCUUGAGUCAGCGCUUGGAUGU----------------UUGAA--------UGUUUGAAUG-GGUUUAGCCAUAGG ....((((((((((......(((((((((.........))).))))))..))))))))))..(((.----------------(((((--------(.(......)-.)))))).)))... ( -30.40) >DroSec_CAF1 19382 103 + 1 CAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGAGAGUGCUCCCCUUGAGUCAGCGCCUCGAUGU----------------UUGAAUGUUUGAAUGUUUGAAUG-GGUUUAGCCAUGGG ....((((((((((.((((..((((..(.......)..))))..))))..))))))).)))...((----------------(..(((((....)))))..))).-((.....))..... ( -28.30) >DroSim_CAF1 19097 95 + 1 CAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGAGAGUGCCCCGCUUGAGUCAGCGCCUCGAUGU----------------UUGGA--------UGUUUGAAUG-GGUUUAGCCAUGGG ...((.((((((((......(((((((((.........)).)))))))..))))))))))(.(((.----------------(((((--------(.(......)-.)))))).))).). ( -29.40) >DroEre_CAF1 21095 95 + 1 CAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGAGAGUGCUCCGCUUGAGUCAGCGACUCGAUGU----------------UUGGA--------UGUUUGAAUG-GGUUUAGCAGUGGG ....((((((((((......(((((((((.........))).))))))..)))))).))))..(((----------------(.(((--------(.(......)-.))))))))..... ( -27.80) >DroYak_CAF1 21727 112 + 1 CAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGUGAGUGCUCCGCUUAAGUCAGCGACUCGAUGUUUAAAUGUUUGAAUGUUUGGA--------UGUUUGAAUGGGGCUUAGCAAUGGG ....((((((((((......(((((((((.........))).))))))..)))))).))))...(((((((((((..(....)..))--------)))))))))....((((....)))) ( -33.70) >consensus CAAAGAGUGCUGAUGAAGGAAAGCGGGCAAGAAGAGAGUGCUCCGCUUGAGUCAGCGCCUCGAUGU________________UUGGA________UGUUUGAAUG_GGUUUAGCCAUGGG ....((((((((((......(((((((((.........))).))))))..))))))).)))........................................................... (-21.76 = -22.00 + 0.24)

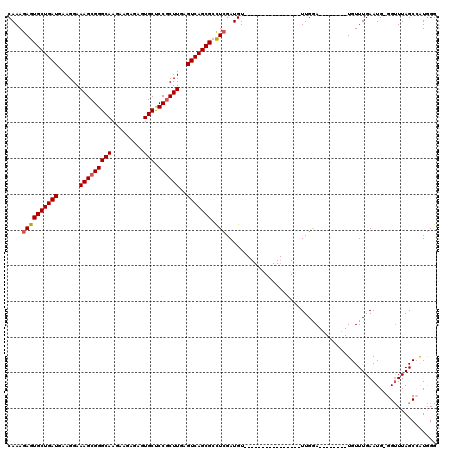
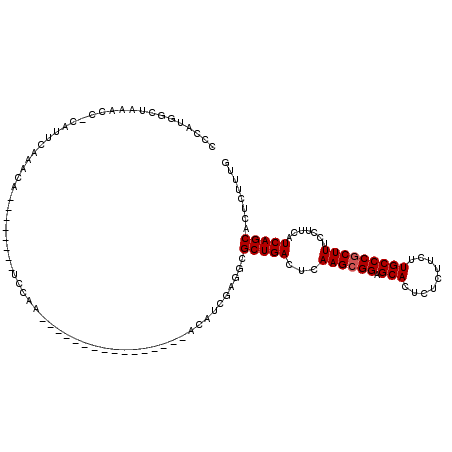
| Location | 13,048,960 – 13,049,055 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.33 |
| Mean single sequence MFE | -21.69 |
| Consensus MFE | -15.78 |
| Energy contribution | -15.98 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13048960 95 - 20766785 CCUAUGGCUAAACC-CAUUCAAACA--------UUCAA----------------ACAUCCAAGCGCUGACUCAAGCGGAGCACUCUCUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUG ...((((......)-))).......--------.....----------------.......((.(((((...((((((.(((.........))))))))).......))))).))..... ( -19.00) >DroSec_CAF1 19382 103 - 1 CCCAUGGCUAAACC-CAUUCAAACAUUCAAACAUUCAA----------------ACAUCGAGGCGCUGACUCAAGGGGAGCACUCUCUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUG ...((((......)-)))....................----------------.((..(((..(((((...((((..(((...............)))..))))..))))).)))..)) ( -23.36) >DroSim_CAF1 19097 95 - 1 CCCAUGGCUAAACC-CAUUCAAACA--------UCCAA----------------ACAUCGAGGCGCUGACUCAAGCGGGGCACUCUCUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUG ...((((......)-))).......--------.....----------------.((..(((..(((((...(((((((.((.........))))))))).......))))).)))..)) ( -22.20) >DroEre_CAF1 21095 95 - 1 CCCACUGCUAAACC-CAUUCAAACA--------UCCAA----------------ACAUCGAGUCGCUGACUCAAGCGGAGCACUCUCUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUG ..............-..........--------.....----------------.((..((((.(((((...((((((.(((.........))))))))).......)))))))))..)) ( -21.60) >DroYak_CAF1 21727 112 - 1 CCCAUUGCUAAGCCCCAUUCAAACA--------UCCAAACAUUCAAACAUUUAAACAUCGAGUCGCUGACUUAAGCGGAGCACUCACUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUG .........................--------......................((..((((.(((((...((((((.(((.........))))))))).......)))))))))..)) ( -22.30) >consensus CCCAUGGCUAAACC_CAUUCAAACA________UCCAA________________ACAUCGAGGCGCUGACUCAAGCGGAGCACUCUCUUCUUGCCCGCUUUCCUUCAUCAGCACUCUUUG ................................................................(((((...((((((.(((.........))))))))).......)))))........ (-15.78 = -15.98 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:15 2006