
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,047,388 – 13,047,516 |
| Length | 128 |
| Max. P | 0.931200 |

| Location | 13,047,388 – 13,047,482 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 79.10 |
| Mean single sequence MFE | -29.27 |
| Consensus MFE | -18.68 |
| Energy contribution | -19.57 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13047388 94 - 20766785 AAAAGAGUUUGGG----------------GGGGAAAACAAAGGAAGCCGGCUGGCGUUUGGCGGCGCACGCCAAGCCAAACACGACCGAGUUUAUCGAACUCAUAUUGAA ....((((((((.----------------.(......).....(((((((.(.(.((((((((((....)))..))))))).).)))).)))).))))))))........ ( -31.10) >DroSec_CAF1 17817 94 - 1 AAAAGAGUUUGGG----------------GGGAAAAACAAAGGAAGCCGGCUGGCGGUUGGCGGCGCACGCCAAGCCAAACACGAGCGAGUUUAUCGAACUCAUAUUGAA ....((((((((.----------------.(......).....((((..(((.(.(.((((((((....)))..))))).).).)))..)))).))))))))........ ( -27.90) >DroSim_CAF1 17557 96 - 1 AAAAGAGUUUGGGGG--------------GGGAAAAACAAAGGAAGCCGGCUGGCGGUUGGCGGCGCACGCCAAGCCAAACACGAGCGAGUUUAUCGAACUCAUAUUGAA ....((((((((...--------------.(......).....((((..(((.(.(.((((((((....)))..))))).).).)))..)))).))))))))........ ( -27.90) >DroEre_CAF1 19543 103 - 1 GAAAGAGUUUGGUGGGGAGCGAGGGGGAGGGGGGAAACAAAGAAAGGCGGC-------UGGCGGCGCACGCCAAGCCAAACACGAGCGAGUUUAUCGAACUCAUAUUGAA ....((((((((((((..((..(.........(....)..........(((-------(((((.....)))).)))).....)..))...))))))))))))........ ( -33.00) >DroYak_CAF1 20122 96 - 1 AAAAGAGUUUGGGGGGGA--------------AAAAACAAAGGAAGGCGGCUGCCGGCUGACGGCGCACGCCAAGCCAAACACGAGCGAGUUUAUCGAACUCAUAUUGAA ....((((((((......--------------.........((..((((((.((((.....)))))).))))...))((((.(....).)))).))))))))........ ( -30.70) >DroPer_CAF1 21921 96 - 1 AAAACAAUAGGAUGGGGA--------------AGAGGAAGGGGCACGUGGUCGGAGACUGUCGGCGCACGCCAAGCCAAACGCGAGCGGGUUUAUCGAACUCAUAUUGAA ....(((((...((((..--------------.((......(((..((((((((......))))).))))))(((((...(....)..))))).))...))))))))).. ( -25.00) >consensus AAAAGAGUUUGGGGG______________GGGAAAAACAAAGGAAGCCGGCUGGCGGUUGGCGGCGCACGCCAAGCCAAACACGAGCGAGUUUAUCGAACUCAUAUUGAA ......((((((.................................((((((.....))))))(((....)))...)))))).(((..((((((...))))))...))).. (-18.68 = -19.57 + 0.89)

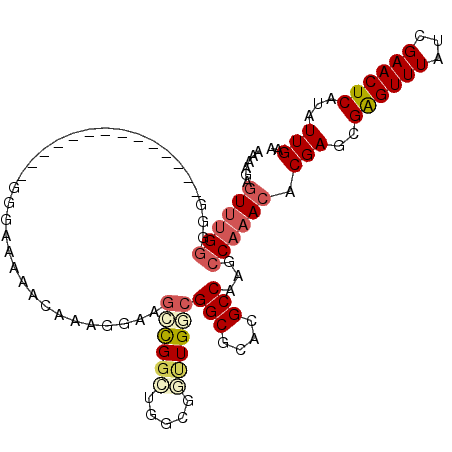
| Location | 13,047,425 – 13,047,516 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 71.58 |
| Mean single sequence MFE | -37.35 |
| Consensus MFE | -12.59 |
| Energy contribution | -14.53 |
| Covariance contribution | 1.95 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13047425 91 - 20766785 CUACCCAGC-----CACUCCCUUCCUGGCCCCAAAAUCCAAAAGAGUUUGGG----------------GGGGAAAACAAAGGAAGCCGGCUGGCGUUUGGCGGCGCACGCCA ..(((((((-----(.((.(((((((..(((((((.((.....)).))))))----------------)))))......))).))..)))))).)).(((((.....))))) ( -37.10) >DroSec_CAF1 17854 91 - 1 CUACACAGC-----CACUCCCUUCCUGGCCCCAAAAUCCAAAAGAGUUUGGG----------------GGGAAAAACAAAGGAAGCCGGCUGGCGGUUGGCGGCGCACGCCA .......((-----((((..((((((..(((((((.((.....)).))))))----------------)(......)..))))))..)).))))...(((((.....))))) ( -33.80) >DroSim_CAF1 17594 93 - 1 CUACCCAGC-----CACUCCCUUCCUGGCCCCAAAAUCCAAAAGAGUUUGGGGG--------------GGGAAAAACAAAGGAAGCCGGCUGGCGGUUGGCGGCGCACGCCA ...((((((-----(.....((((((..((((....((((((....)))))).)--------------)))........))))))..)))))).)..(((((.....))))) ( -37.80) >DroEre_CAF1 19580 100 - 1 CUACCCAGC-----CACUCCCUUCCUGGCCCCAAAAUCCGAAAGAGUUUGGUGGGGAGCGAGGGGGAGGGGGGAAACAAAGAAAGGCGGC-------UGGCGGCGCACGCCA ....(((((-----(.((((((((((.(((((.(((..(....)..)))....))).)).)))))).))))(....)..........)))-------))).(((....))). ( -47.30) >DroYak_CAF1 20159 93 - 1 CUACCCAGC-----CACUCCCUGCCUGGCCCCAAAAUCCAAAAGAGUUUGGGGGGGA--------------AAAAACAAAGGAAGGCGGCUGCCGGCUGACGGCGCACGCCA ...(((.((-----((.........))))((((((.((.....)).)))))))))..--------------.............((((((.((((.....)))))).)))). ( -35.70) >DroPer_CAF1 21958 88 - 1 GCAG-CAGGCAUGGCAUUCCUUUCCC--UCCC-------AAAACAAUAGGAUGGGGA--------------AGAGGAAGGGGCACGUGGUCGGAGACUGUCGGCGCACGCCA ((..-...)).((((..(((((((((--((((-------....(....)....))).--------------)).))))))))...((((((((......))))).))))))) ( -32.40) >consensus CUACCCAGC_____CACUCCCUUCCUGGCCCCAAAAUCCAAAAGAGUUUGGGGG______________GGGAAAAACAAAGGAAGCCGGCUGGCGGUUGGCGGCGCACGCCA ............................(((((((.((.....)).)))))))...............................((((((.....))))))(((....))). (-12.59 = -14.53 + 1.95)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:12 2006