
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,039,190 – 13,039,293 |
| Length | 103 |
| Max. P | 0.690896 |

| Location | 13,039,190 – 13,039,293 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.80 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -16.01 |
| Energy contribution | -15.23 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13039190 103 + 20766785 GUAAAGCAACUCUCU-----------UG------UACCUCUCGCUCGGCCUCUUCAAUGGCUUUCUUGAGUCUGUGCUGCUCCAAAAUGGCCGCUCGCCGUGCCAUUUGCUCCUCCUUUU ....(((........-----------.(------(((..((((...((((........))))....))))...)))).......(((((((((.....)).))))))))))......... ( -24.00) >DroSec_CAF1 9639 100 + 1 GUA---CAACUCUCU-----------UG------UACCUCUCGCUCGGCCUCUUCAAUGGCUUUCUUGAGUCUGUGCUGCUCCAAAAUGGCCGCUCGCCGUGCCAUUUGCUCCUCCUUUU (((---(((.....)-----------))------))).....((.((((.........(((((....))))).(.((.(((.......))).)).))))).))................. ( -22.90) >DroSim_CAF1 9385 100 + 1 GUA---CAACUCUCU-----------UG------UACCUCUCGCUCGGCCUCUUCAAUGGCUUUCUUGAGUCUGUGCUGCUCCAAAAUGGCCGCUCUCCGUGCCAUUUGCUCCUCCUUUU (((---((((((...-----------.(------(.......))..((((........)))).....)))).))))).......(((((((((.....)).)))))))............ ( -21.80) >DroWil_CAF1 12680 105 + 1 -UA---AAGGGCUCC-----------UGAUAAACAACCUCUCUUUCGGCUUCUUCAAUGGCUUUUUUGAGUCUAUGCUGCUCUAAUAUGGCCGCUCGGCGCGCCAUUUGUUCUUCCUUUU -..---.(((((..(-----------(((...............))))..........(((((....)))))......)))))...((((((((...))).))))).............. ( -20.06) >DroAna_CAF1 10040 96 + 1 AUG---GAG----UG-----------AG------UACCUCUCGUUCAGCCUCUUCGAUGGCUUUCUUGAGUCUGUGCUGCUCCAGAAUGGCUGCUCGUCGCGCCAUCUGCUCUUCCUUCU ..(---(((----(.-----------((------(((..((((...((((........))))....))))...))))))))))(((.((((.((.....)))))))))............ ( -32.00) >DroPer_CAF1 13621 109 + 1 --C---CAACUAUAUUCUGUAGGUGGCU------UACCUCUCGUUCGGCCUCUUCGAUGGCUUUCUUGAGUCUAUGCUGUUCCAAUAUGGCCGCUCGUCGCGCCAUUUGAUCCUCCUUCU --.---...((((.....))))(..((.------.....((((...((((........))))....)))).....))..)..(((.(((((.((.....))))))))))........... ( -23.20) >consensus GUA___CAACUCUCU___________UG______UACCUCUCGCUCGGCCUCUUCAAUGGCUUUCUUGAGUCUGUGCUGCUCCAAAAUGGCCGCUCGCCGCGCCAUUUGCUCCUCCUUUU .......................................((((...((((........))))....))))............(((.(((((.((.....))))))))))........... (-16.01 = -15.23 + -0.77)

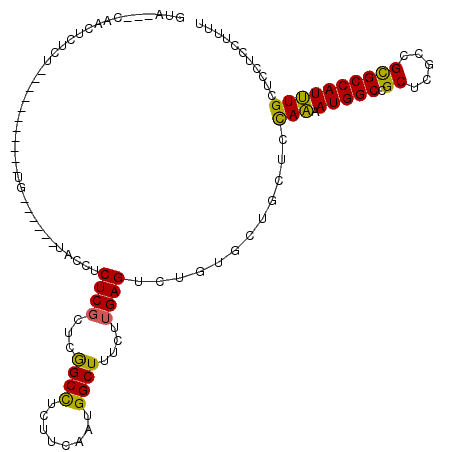
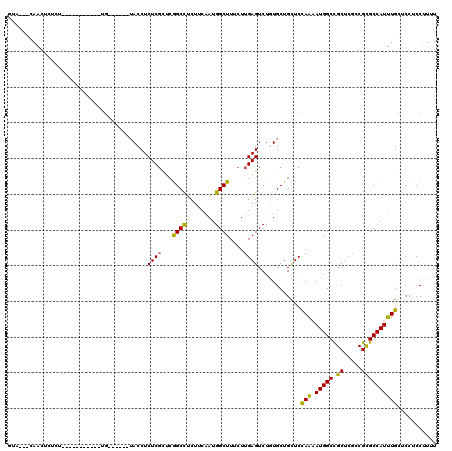
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:06 2006