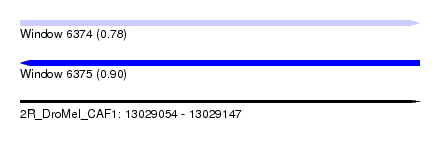
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,029,054 – 13,029,147 |
| Length | 93 |
| Max. P | 0.901535 |

| Location | 13,029,054 – 13,029,147 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -21.75 |
| Consensus MFE | -9.45 |
| Energy contribution | -8.83 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13029054 93 + 20766785 -AUUUCCCCAGUUGGGG--AUAUCGAUUAUUGCCAAAUUUGUGUAAACAUCGAUGGUAUCGCUG------------UACCGAGACAACAAUAAUAGAAAUAAA--UUAAG -...(((((....))))--).((((((..((((((....)).))))..))))))(((((....)------------)))).......................--..... ( -20.20) >DroSec_CAF1 12283 93 + 1 -UUUUCCCCAGUUGGGG--CUAUCGAUUAUUGCCAAAUUUGUGUAAACAUCGAUUGUAUCGCUG------------UACCGAGACAACAAUAAUAGAAAUAAA--UUAAG -....((((....))))--((((((((..((((((....)).))))..)))))((((.(((...------------...))).))))......))).......--..... ( -17.00) >DroSim_CAF1 8683 93 + 1 -AUUUCUCCAGUUGGAG--CUAUCGAUUAUUGCCAAAUUUGUGUAAACAUCGAUUGUAUCGCUG------------UACCGAGACAACAAUAAUAGAAAUAAA--UUAAG -...((((((((.((.(--(.((((((..((((((....)).))))..)))))).)).))))))------------....))))...................--..... ( -15.70) >DroAna_CAF1 8661 107 + 1 UGCGUCCAGAGAUGGAGAAACAUCGAUAUUUUUAAACAUUUUAAAAACAUCGAUUA-AUCGAUGUUUCUCCACCUCUAUCGCAAUAACG--AAUAAAAAUAAAAAUCUGC ((((...((((.(((((((((((((((.(((((((.....))))))).)))((...-.)))))))))))))).))))..))))......--................... ( -34.10) >consensus _AUUUCCCCAGUUGGAG__AUAUCGAUUAUUGCCAAAUUUGUGUAAACAUCGAUUGUAUCGCUG____________UACCGAGACAACAAUAAUAGAAAUAAA__UUAAG .....((((....))))....((((((..((((((....)).))))..))))))........................................................ ( -9.45 = -8.83 + -0.62)

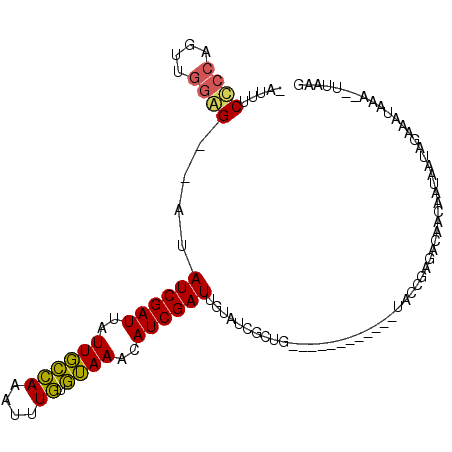
| Location | 13,029,054 – 13,029,147 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -9.07 |
| Energy contribution | -9.82 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13029054 93 - 20766785 CUUAA--UUUAUUUCUAUUAUUGUUGUCUCGGUA------------CAGCGAUACCAUCGAUGUUUACACAAAUUUGGCAAUAAUCGAUAU--CCCCAACUGGGGAAAU- .....--...........(((((((((......)------------))))))))..(((((((((..((......))..))).)))))).(--((((....)))))...- ( -21.60) >DroSec_CAF1 12283 93 - 1 CUUAA--UUUAUUUCUAUUAUUGUUGUCUCGGUA------------CAGCGAUACAAUCGAUGUUUACACAAAUUUGGCAAUAAUCGAUAG--CCCCAACUGGGGAAAA- .....--.......((((((((((((((...(((------------((.((((...)))).)))....))......))))))))).)))))--((((....))))....- ( -20.70) >DroSim_CAF1 8683 93 - 1 CUUAA--UUUAUUUCUAUUAUUGUUGUCUCGGUA------------CAGCGAUACAAUCGAUGUUUACACAAAUUUGGCAAUAAUCGAUAG--CUCCAACUGGAGAAAU- .....--.......((((((((((((((...(((------------((.((((...)))).)))....))......))))))))).)))))--((((....))))....- ( -18.60) >DroAna_CAF1 8661 107 - 1 GCAGAUUUUUAUUUUUAUU--CGUUAUUGCGAUAGAGGUGGAGAAACAUCGAU-UAAUCGAUGUUUUUAAAAUGUUUAAAAAUAUCGAUGUUUCUCCAUCUCUGGACGCA ((................(--(((....))))((((((((((((((((((((.-...))(((((((((((.....)))))))))))))))))))))))))))))...)). ( -43.20) >consensus CUUAA__UUUAUUUCUAUUAUUGUUGUCUCGGUA____________CAGCGAUACAAUCGAUGUUUACACAAAUUUGGCAAUAAUCGAUAG__CCCCAACUGGGGAAAU_ ..................((((((((....................))))))))..((((((.......((....))......))))))....((((....))))..... ( -9.07 = -9.82 + 0.75)

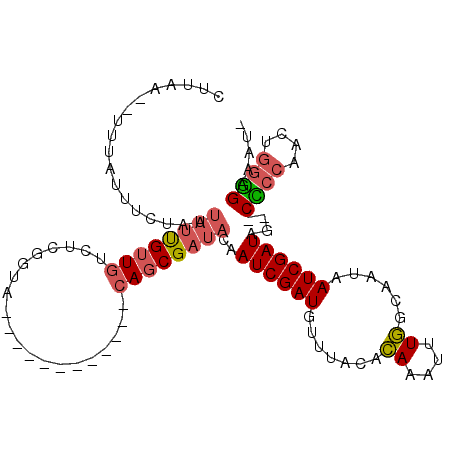
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:08:03 2006