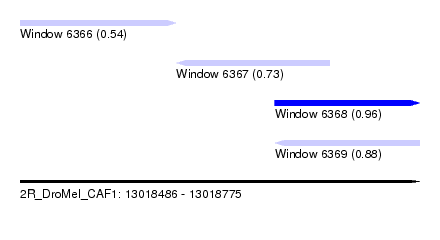
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,018,486 – 13,018,775 |
| Length | 289 |
| Max. P | 0.962812 |

| Location | 13,018,486 – 13,018,599 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -20.97 |
| Energy contribution | -20.37 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.539351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
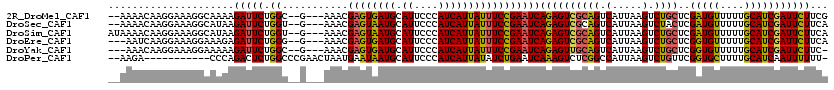
>2R_DroMel_CAF1 13018486 113 + 20766785 --AAAACAAGGAAAGGCAAAAGAUUCUGGC--G---AAACGAGUGAUGCAUUCCCAUCAUUAUUUCCGAAUCAGAGUCGCAGUCAUUAAGUCUGCUCGAUGUUUUUGCAUCGAUUCUUCG --......(((((........(((((((.(--(---.....(((((((......))))))).....))...)))))))((((.(.....).))))(((((((....)))))))))))).. ( -32.10) >DroSec_CAF1 32144 113 + 1 --AAAACAAGGAAAGGCAUAAGAUUCUGGU--G---AAACGAGUAAUGCAUUCCCAUCAUUAUUUCCGAAUCAGAGUCGCAGUCAUUAAGUCUACUCGAUGUUUUUGCAUCGAUUCUUCA --......(((((.(((....(((((((((--.---....((((((((.((....))))))))))....)))))))))...)))...........(((((((....)))))))))))).. ( -28.10) >DroSim_CAF1 28502 115 + 1 AUAAAACAAGGAAAGGCAUAAGAUUCUGGU--G---AAACGAGUAAUGCAUUCCCAUCAUUAUUUCCGAAUCAGAGUCGCAGUCAUUAAGUCUGCUCGAUGUUUUUGCAUCGAUUCUUCA ........(((((........(((((((((--.---....((((((((.((....))))))))))....)))))))))((((.(.....).))))(((((((....)))))))))))).. ( -31.00) >DroEre_CAF1 28007 112 + 1 ---AAUCAAGGAAAGGAAAGAGAUUCUGGG--G---AAACGAGUGAUGCAUUCCCAUCAUUAUUUCCGAAUCAGAGUCGCAGUCAUUAAGUCUGCUCGGUGUUUUUGCAUCGAUUCUUCA ---.......(((.(((....(((((((((--(---(((..(((((((......))))))).)))))...))))))))((((.(.....).))))(((((((....))))))))))))). ( -35.20) >DroYak_CAF1 28839 111 + 1 ---AAACAAGGAAAGGAAAAAGAUUCUGGC--G---AAACGAGUGAUGCAUUCCCAUCAUUAUUUCCGAAUCAGAGUUGCAGUCAUUAAGUCUGCUCGGUGUUUUUGCAUCGAUUCUUC- ---..........(((((...(((((((.(--(---.....(((((((......))))))).....))...)))))))((((.(.....).))))(((((((....)))))))))))).- ( -28.70) >DroPer_CAF1 27087 106 + 1 --AAGA-----------CCCAGACUCUGGCCCGAACUAAUGAAUAAUGCAUUCCCAUCAUUAUAUCUGAAUCAAAGUCUCGGCCAUUAAGUCUGUUCGGUGCUUUUGCAUCAAUUUUUU- --..((-----------..((((((.(((((.(((((..(((((((((.((....)))))))).......))).))).)))))))...)))))).))(((((....)))))........- ( -25.11) >consensus __AAAACAAGGAAAGGCAAAAGAUUCUGGC__G___AAACGAGUAAUGCAUUCCCAUCAUUAUUUCCGAAUCAGAGUCGCAGUCAUUAAGUCUGCUCGAUGUUUUUGCAUCGAUUCUUCA .....................(((((.((...........((((((((.((....)))))))))))))))))((((((((((.(.....).))))..(((((....)))))))))))... (-20.97 = -20.37 + -0.61)
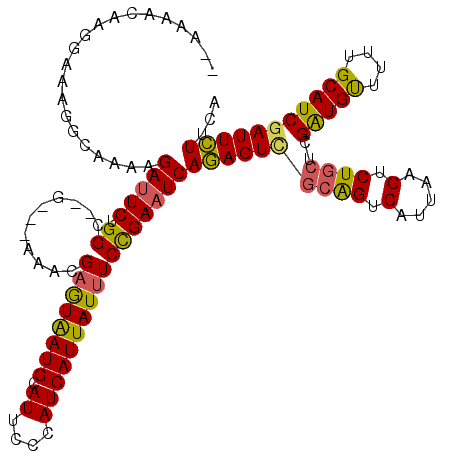
| Location | 13,018,599 – 13,018,710 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.08 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -15.60 |
| Energy contribution | -17.78 |
| Covariance contribution | 2.18 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13018599 111 - 20766785 GAAUAUAAAGCCAGUUUGUUCGCUAUCGGGAACAGGAAUGGGAACAAUGCUGGUGGGUAAUCGAUGAGAAGAGAGCUCUCA--GCGAAGACUAGAGAGUGAAGACUCUGGACC .........((((((((((((.(((((........).)))))))))).)))))).(((..(((.(((((.......)))))--.)))...((((((........))))))))) ( -32.80) >DroSec_CAF1 32257 105 - 1 GAAAAUAAAGCCAGUUUGUUGGCCAUCAG------GAACGGGAACAAUGCUGGUGGGUAAUCGAUGAGAAGAGAGCUCUCA--GCGAAGACUGGAGAGUGAAGACUCCGGACC ..........(((((((.(((.(((((((------.(..(....)..).))))))).)))(((.(((((.......)))))--.)))))))))).((((....))))...... ( -32.10) >DroSim_CAF1 28617 105 - 1 GAAAAUAAAGCCAGUUUGUUGGCCAUCAG------GAACGGGAACAAUGCUGGUGGGUAAUCGAUGAGAAGAGAGCUCUCA--GCGAAGACUGGAGAGUGAAGACUCUGGACC ..........(((((((.(((.(((((((------.(..(....)..).))))))).)))(((.(((((.......)))))--.))))))))))(((((....)))))..... ( -33.80) >DroEre_CAF1 28119 92 - 1 -UAUAUAAAGCCAGUU----CGGCAUCGG------GAACGGGAACAACGCUGGUGGGUAAUCGAUGAGAAGAGAGCUCACU--GCGAAGACUGG------AAGAC--UGGACC -.........((((((----((((.(((.------...)))......(((.(((((((..((........))..)))))))--)))..).))).------..)))--)))... ( -26.80) >DroYak_CAF1 28950 96 - 1 -AAAAUAAAGCCAGUUUGUUUGGCAUCGG------GAAUGGGAACACUGCUGGUGGGUAAUCGAUGAGAAGAGAGCUCAGAGAGUGAAGACUCU------GAACC--UGGA-- -........(((((.....))))).((((------(..((((..(.((.((.((.(.....).)).)).)).)..)))).(((((....)))))------...))--))).-- ( -24.50) >consensus GAAAAUAAAGCCAGUUUGUUCGCCAUCGG______GAACGGGAACAAUGCUGGUGGGUAAUCGAUGAGAAGAGAGCUCUCA__GCGAAGACUGGAGAGUGAAGACUCUGGACC .........((((((((((((....((........))....)))))).))))))((((..((........))..))))............((((((........))))))... (-15.60 = -17.78 + 2.18)


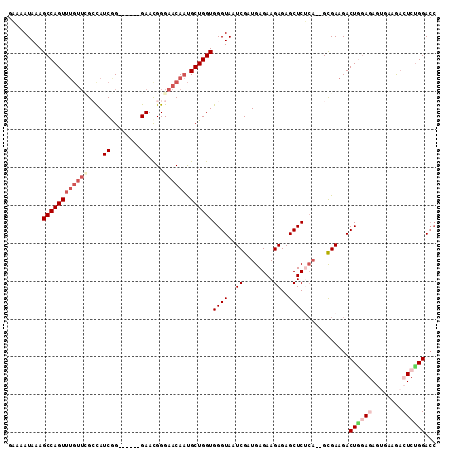
| Location | 13,018,670 – 13,018,775 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 75.24 |
| Mean single sequence MFE | -10.20 |
| Consensus MFE | -8.12 |
| Energy contribution | -9.12 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
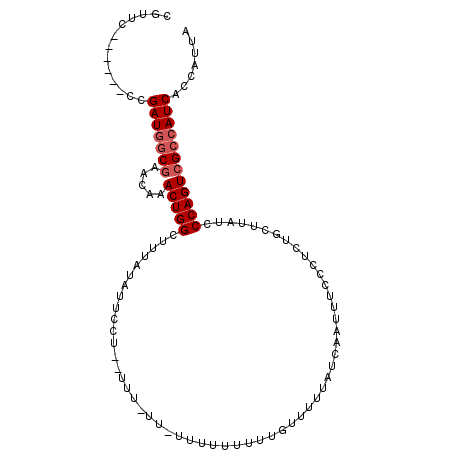
>2R_DroMel_CAF1 13018670 105 + 20766785 CAUUCCUGUUCCCGAUAGCGAACAAACUGGCUUUAUAUUCCUUAUUU-UUUUUUUUUUUUGUUUUUAUCAAUUUCCCUGUGCUUAUCCCAGUCGCCAUCACCAUUA .............(((.((((.......(((................-................................)))........)))).)))....... ( -8.87) >DroSec_CAF1 32328 100 + 1 CGUUC------CUGAUGGCCAACAAACUGGCUUUAUUUUCCUCCUUUUUUCUUUUUUUUUGUUUUUAUCAAUUUCCCUCUGCUUAUCCCAGUCGCCAUCACCAUUA .....------.(((((((......(((((.........................................................))))).)))))))...... ( -11.12) >DroEre_CAF1 28182 79 + 1 CGUUC------CCGAUGCCG----AACUGGCUUUAUAUA-----------------UAUUUUUUUUAUCAAUUUCCCUCUGCUUAUCCCAGUCGCCAUCACCAUUA .....------..((((.((----(.((((.........-----------------...............................))))))).))))....... ( -10.60) >consensus CGUUC______CCGAUGGCGAACAAACUGGCUUUAUAUUCCU__UUU_UU_UUUUUUUUUGUUUUUAUCAAUUUCCCUCUGCUUAUCCCAGUCGCCAUCACCAUUA .............(((((((.....(((((.........................................................))))))))))))....... ( -8.12 = -9.12 + 1.00)

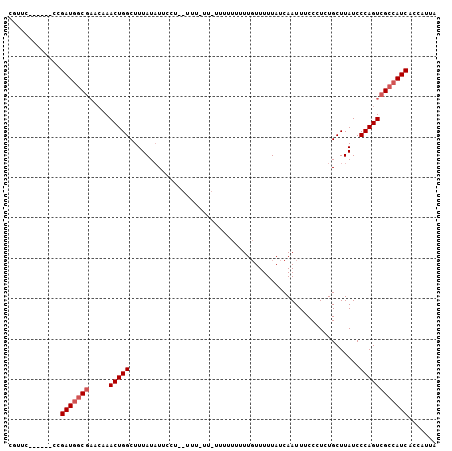
| Location | 13,018,670 – 13,018,775 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 75.24 |
| Mean single sequence MFE | -14.50 |
| Consensus MFE | -11.06 |
| Energy contribution | -10.95 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
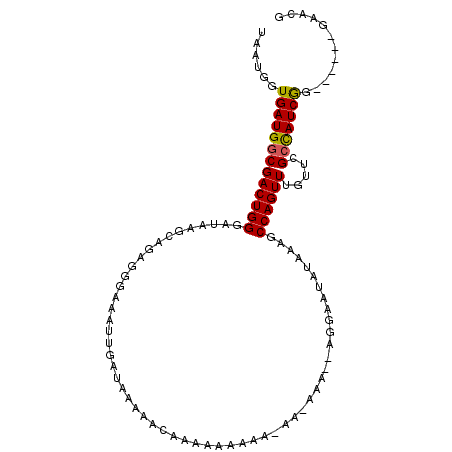
>2R_DroMel_CAF1 13018670 105 - 20766785 UAAUGGUGAUGGCGACUGGGAUAAGCACAGGGAAAUUGAUAAAAACAAAAAAAAAAAA-AAAUAAGGAAUAUAAAGCCAGUUUGUUCGCUAUCGGGAACAGGAAUG .......((((((((.....((((((.(((.....)))....................-......((.........)).))))))))))))))(....)....... ( -15.20) >DroSec_CAF1 32328 100 - 1 UAAUGGUGAUGGCGACUGGGAUAAGCAGAGGGAAAUUGAUAAAAACAAAAAAAAAGAAAAAAGGAGGAAAAUAAAGCCAGUUUGUUGGCCAUCAG------GAACG ......((((((((((((((.....).........(((.......)))............................)))))).....))))))).------..... ( -14.50) >DroEre_CAF1 28182 79 - 1 UAAUGGUGAUGGCGACUGGGAUAAGCAGAGGGAAAUUGAUAAAAAAAAUA-----------------UAUAUAAAGCCAGUU----CGGCAUCGG------GAACG .......((((.(((((((...............................-----------------.........)))).)----)).))))(.------...). ( -13.80) >consensus UAAUGGUGAUGGCGACUGGGAUAAGCAGAGGGAAAUUGAUAAAAACAAAAAAAAA_AA_AAA__AGGAAUAUAAAGCCAGUUUGUUCGCCAUCGG______GAACG ......(((((((((((((.........................................................)))))).....)))))))............ (-11.06 = -10.95 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:57 2006