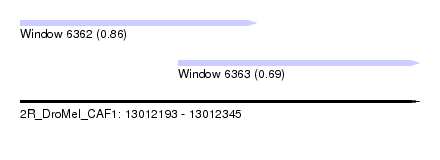
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,012,193 – 13,012,345 |
| Length | 152 |
| Max. P | 0.855672 |

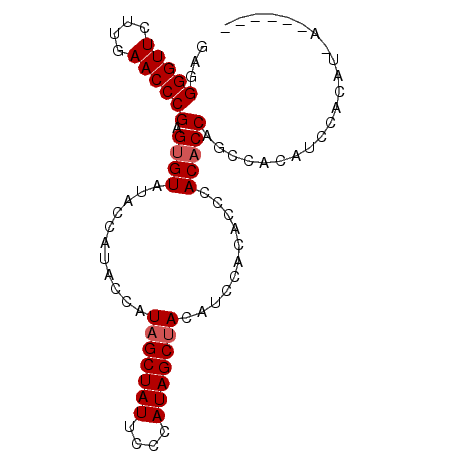
| Location | 13,012,193 – 13,012,283 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 85.23 |
| Mean single sequence MFE | -16.21 |
| Consensus MFE | -12.82 |
| Energy contribution | -13.48 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13012193 90 + 20766785 GAGGGGUUCUUGAACCCGAGUGUAUACCAUACCAUUGCUAUUCCCAUAGCUACAUCCACACCCACACCAGCCACAUCCACAUUACAUCCA ...(((((....)))))(.((((.............(((((....))))).............)))))...................... ( -13.87) >DroSec_CAF1 25974 76 + 1 GAGGGGUUCUUGAACCCGAGUGUGUACCAUAACAUAGCUAUUCCCAUAGCUACAUCCACACCCACACCAGCCACAU-------------- ...(((((....)))))(.(((((..........(((((((....)))))))..........))))))........-------------- ( -18.95) >DroSim_CAF1 22318 84 + 1 GAGGGGUUCUUGAACCCGAGAGUAUACCAUACCAUAGCUAUUCCCAUAGCUACAUGCACACCCACACCAGCCACAUCCACAUCA------ ..(((.((((((....))))))............(((((((....)))))))........))).....................------ ( -15.80) >consensus GAGGGGUUCUUGAACCCGAGUGUAUACCAUACCAUAGCUAUUCCCAUAGCUACAUCCACACCCACACCAGCCACAUCCACAU_A______ ...(((((....)))))(.((((...........(((((((....)))))))...........)))))...................... (-12.82 = -13.48 + 0.67)


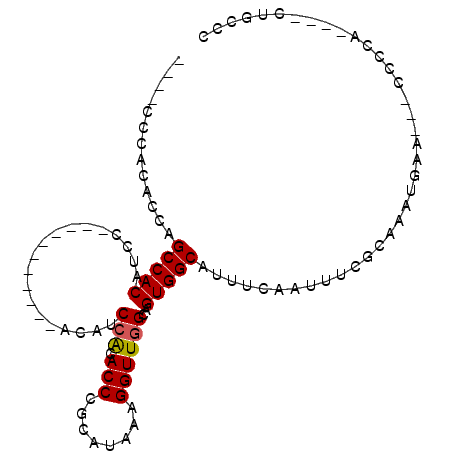
| Location | 13,012,253 – 13,012,345 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -16.32 |
| Consensus MFE | -9.10 |
| Energy contribution | -9.13 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13012253 92 + 20766785 ----CCCACACCAGCCACAUCCACAUUACAUCCACAUCCACACCCGCAUAAAGGUUGGCAGUGGCAUUUCAAUUUCGCAAAUGAA---CCCCA----CUGCCC ----.....................................(((........))).((((((((...((((..........))))---..)))----))))). ( -16.30) >DroSec_CAF1 26034 75 + 1 ----CCCACACCAGCCACAU-----------------CCACACCCGCAUAAAGGUUGGCAGUGGCAUUUCAAUUUCGCAAAUGAA---CCCCA----CUGCCC ----........((((....-----------------...............))))((((((((...((((..........))))---..)))----))))). ( -16.71) >DroSim_CAF1 22378 86 + 1 ----CCCACACCAGCCACAUCCACAUCA------CAUCCACACCCGCAUAAAGGUUGGCAGUGGCAUUUCAAUUUCGCAAAUGAA---CCCCA----CUGCCC ----........................------.......(((........))).((((((((...((((..........))))---..)))----))))). ( -16.30) >DroEre_CAF1 21962 81 + 1 ----CCCACACUAGCCACAUCC-----------ACAUCCACACCCGCAUAAAGGUUGGCAGUGGCAUUUCAAUUUCGCAAAUGAA---CCCCG----CUGCCC ----........((((......-----------...................))))((((((((...((((..........))))---..)))----))))). ( -15.91) >DroYak_CAF1 22666 89 + 1 ACACCCCACACCAGCCACAUUC-----------ACAUCCACACCCGCAUAAAGGUUGGCAGUGGCAUUUCAAUUUCGCAAAUGAA---CCCCACUCACUGCCC .....((((.((((((......-----------...................))))))..))))............(((..(((.---......))).))).. ( -13.91) >DroAna_CAF1 29145 83 + 1 -----------CUGCCACAUCCGCAC-----CCGCACCAGCACCCGCAUAAAGGUUGGCAGUGGCAUUUCAAUUUCGCAAAUGAAUAGCCCCC----CUUCCC -----------((((((...((((..-----..))....((....)).....)).)))))).(((..((((..........))))..)))...----...... ( -18.80) >consensus ____CCCACACCAGCCACAUCC___________ACAUCCACACCCGCAUAAAGGUUGGCAGUGGCAUUUCAAUUUCGCAAAUGAA___CCCCA____CUGCCC .............(((((...................(((.(((........))))))..)))))...................................... ( -9.10 = -9.13 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:52 2006