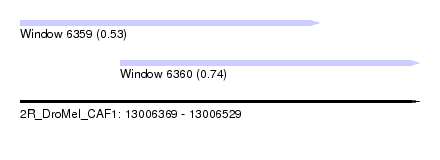
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 13,006,369 – 13,006,529 |
| Length | 160 |
| Max. P | 0.740625 |

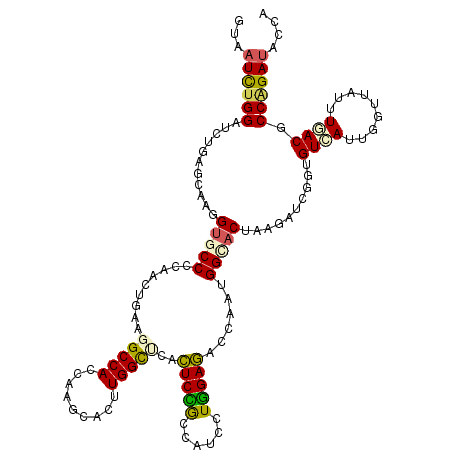
| Location | 13,006,369 – 13,006,489 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -36.62 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.48 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.527707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13006369 120 + 20766785 GUAAUCUGGACCUAAGCAAGGUUCCCCAACUGAAGGCCACUUGGCACUUGGCUAACUCCGCCAUUCUGGAGACCAAUGGCACUAAGGUCGGUGUCAUAGGUUAUUUGACUCCAGAUACCA .......((((((.....))))))........((((((....))).)))(((.......)))..((((((((((.((((((((......)))))))).))).......)))))))..... ( -38.51) >DroPse_CAF1 14748 120 + 1 GCAAUCUGGAUAUGAGGAAAGUGCCCGAGUUGGCUGCCACCAAACAUUUGGUUCCCUCUACGAUCCUGGAGACCCAGGGCACCAAGAUAGGUGUGAUUGGCUAUUUGACGCCAGAUACUA ...((((((....((((.....(((......)))....(((((....)))))..))))..(((((((((....)))).(((((......))))))))))...........)))))).... ( -36.40) >DroGri_CAF1 19469 120 + 1 GUAAUUUGGAUCUUACUAAUGAGCCCGAAAUGGCUGCCACUAAGCAUUUGGCCAAUUCGACAAUAAUUGAAACAAAUGGUACUAAAAUUGCUGUAAUCGGAUAUGUGACGCCAGAUACGA (((.(((((...((((...(((...((((.(((((((......))....))))).))))................((((((.......))))))..))).....))))..)))))))).. ( -22.70) >DroEre_CAF1 16182 120 + 1 GCAAUCUGGACCUUAGCAAGGUGCCCCAACUGGAGGCCACCAAGCACUUGGCUCACUCCGCCAUCCUGGAGGCCAAUGGCACUAGUAUCGGUGUCAUAGGUUAUUUGACACCGGAUACCA ((....(((.((((.((..((((((((....)).)).))))..))...((((.......)))).....)))))))...))....((((((((((((.........))))))).))))).. ( -44.20) >DroYak_CAF1 16775 120 + 1 GUAAUCUGGAUCUGAGCAAGGUUCCCCAACUGAAGGCCACCAAGCACUUGGCUCACUCCGCCAUCCUGGAGACCAAUGGCACUAAGAUCGGUGUCAUUGGUUACUUGACCCCGGAUACCA ...((((((...(((((..((....)).....(((((......)).))).)))))(((((......)))))((((((((((((......)))))))))))).........)))))).... ( -39.70) >DroAna_CAF1 23921 120 + 1 GUAAUCUGGAUCUGAGCAAAGUGCCCGAACUCAAGGCCACAAAGCACUUGGCCCACUCUGCCAUCCUGGAGGCUAAUGGCACCAAGAUCGGUGUGAUUGGCUACCUCACUCCCGACACCA ....((.(((..((((((.((((...(....)..(((((.........))))))))).)))......((.(((((((.(((((......))))).))))))).))))).))).))..... ( -38.20) >consensus GUAAUCUGGAUCUGAGCAAGGUGCCCCAACUGAAGGCCACCAAGCACUUGGCUCACUCCGCCAUCCUGGAGACCAAUGGCACUAAGAUCGGUGUCAUUGGUUAUUUGACGCCAGAUACCA ...((((((...........(((((.........(((((.........)))))..(((((......)))))......)))))..........((((.........)))).)))))).... (-21.70 = -21.48 + -0.22)


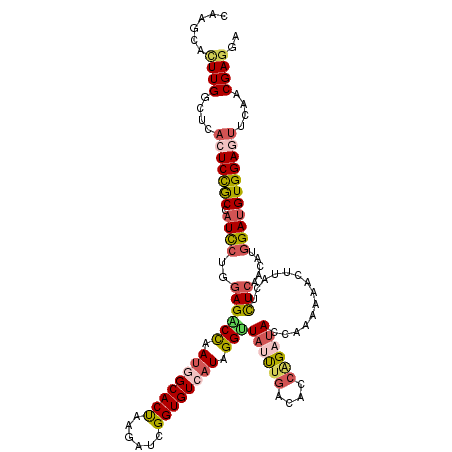
| Location | 13,006,409 – 13,006,529 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.56 |
| Mean single sequence MFE | -37.25 |
| Consensus MFE | -27.22 |
| Energy contribution | -28.20 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 13006409 120 + 20766785 UUGGCACUUGGCUAACUCCGCCAUUCUGGAGACCAAUGGCACUAAGGUCGGUGUCAUAGGUUAUUUGACUCCAGAUACCAAAAAACUUACUCUCAACAUGGAUGUGGAGUUCAACGAGGA ......((((...((((((((...((((((((((.((((((((......)))))))).))).......)))))))..(((..................)))..))))))))...)))).. ( -38.68) >DroSim_CAF1 16466 120 + 1 CAAGCACUUGGCUCACUCCGCCAUCCUUGAGACCAAUGGCACUAAGGUCGGUGUCAUAGGUUAUUUGACACCAGAUACCAAAAAACUUACUCUCAACAUGGAUGUGGAGUUCAACGAAGA ......(((.(...(((((((.((((((((((...((((((((......)))))))).(((.(((((....))))))))...........))))))...)))))))))))....).))). ( -37.80) >DroEre_CAF1 16222 120 + 1 CAAGCACUUGGCUCACUCCGCCAUCCUGGAGGCCAAUGGCACUAGUAUCGGUGUCAUAGGUUAUUUGACACCGGAUACCAAAAAGCUGACAUUGAACAUGGAUGUGGAAUUUAACGAGGA ......((((......(((((.((((....(..(((((.((((.((((((((((((.........))))))).))))).....)).)).)))))..)..)))))))))......)))).. ( -35.80) >DroYak_CAF1 16815 120 + 1 CAAGCACUUGGCUCACUCCGCCAUCCUGGAGACCAAUGGCACUAAGAUCGGUGUCAUUGGUUACUUGACCCCGGAUACCAAAAAGCUUACGCUCAACAUGGAUGUGGAGUUUAACGAGGA ......((((....(((((((.(((((((.(((((((((((((......)))))))))))))......(....)...)))...(((....)))......)))))))))))....)))).. ( -42.00) >DroAna_CAF1 23961 120 + 1 AAAGCACUUGGCCCACUCUGCCAUCCUGGAGGCUAAUGGCACCAAGAUCGGUGUGAUUGGCUACCUCACUCCCGACACCAAGAAACUUACUCUCCAUAUGGAUGUGGACUUUAACGAGGA ......((((......((..(.(((((((((((((((.(((((......))))).))))))....((......))................)))))...)))))..))......)))).. ( -34.10) >DroPer_CAF1 14734 120 + 1 CAAACAUUUGGUUCCCUCUACGAUCCUGGAGACCCAGGGCACCAAGAUAGGUGUGAUUGGCUAUUUGACGCCAGAUACUAAACAGCUUACCCUCCAGAGUGAGGUGGAGUUCUACGAGGA ..............((((......(((((....)))))(((((......)))))....(((........)))(((.(((...((.(((((.(....).))))).)).))))))..)))). ( -35.10) >consensus CAAGCACUUGGCUCACUCCGCCAUCCUGGAGACCAAUGGCACUAAGAUCGGUGUCAUAGGUUAUUUGACACCAGAUACCAAAAAACUUACUCUCAACAUGGAUGUGGAGUUCAACGAGGA ......((((....(((((((.((((..((((((.((((((((......)))))))).)))((((((....))))))..............))).....)))))))))))....)))).. (-27.22 = -28.20 + 0.98)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:49 2006