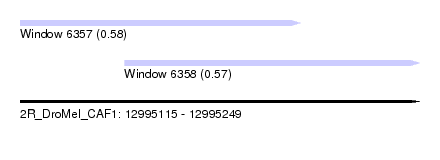
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,995,115 – 12,995,249 |
| Length | 134 |
| Max. P | 0.580250 |

| Location | 12,995,115 – 12,995,209 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 73.45 |
| Mean single sequence MFE | -15.06 |
| Consensus MFE | -6.71 |
| Energy contribution | -7.18 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12995115 94 + 20766785 UCGCCAGAUGCUAUUGUAACUAUUAUUUUAUUGUUUUCCGCA-UUCUCUUACUAUAUUUCAUCAAUAU---ACCUUUCAAUAGUCUCUGGGCAACCAU ..((((((.(((((((...............(((.....)))-.........((((((.....)))))---).....))))))).))).)))...... ( -15.60) >DroVir_CAF1 18183 88 + 1 CAGCCGGAUGCUAUUGUAAGUGCCAAU-CAG--C--UCGUAAAUA-UAUUCCCAUAAUAACUACUU-U---GCUUUUUUGCAGUCACUUGGAAACCAC .....((...(((..((.(.(((.((.-.((--(--..(((..((-(.........)))..)))..-.---)))..)).))).).)).)))...)).. ( -11.30) >DroSec_CAF1 10484 93 + 1 UCACCAGAUGCUAUUGUAACUAUUAAU-UGUUGUUAUCCGCA-UCCUUUUAUUAAGUUUAAUCAACAU---ACAUUUCAAUAGUCUCUGGGCAACCAU ...(((((.(((((((...........-((((((((..(...-............)..))).))))).---......))))))).)))))........ ( -18.93) >DroSim_CAF1 9561 93 + 1 UCACCAGAUGCUAUUGUAACUAUCAAU-UGUUGUUAUCCGCA-UCCUUUUAUUAUAUUUAAUCAACAU---ACACUUCAAUAGUCUCUGGGCAACCAU ...(((((.(((((((...........-((((((((......-...............))).))))).---......))))))).)))))........ ( -17.27) >DroEre_CAF1 9594 93 + 1 UCGCCAGAUGCUAUUGUAACUAUUAAU-UGAUGUUAUCCGCA-UUCGCUCACUAUAUUUUAUGAACCU---GCAUUUGAAUAGUCCCUGGGCAACCAU ....(((((((....(((((.(((...-.))))))))..((.-...))....................---))))))).........(((....))). ( -15.50) >DroYak_CAF1 9739 95 + 1 UCGCCAGAUGCUAUUGUAACUAUUAAC-U-AUCAUAACCGCA-UCAUCUUACCAUAUUUCAUUAAUAUAUAACAUUUGAAUAGUCCCUGGGCAACCAU ..(((.(((((..((((..........-.-...))))..)))-))........(((((.....))))).....................)))...... ( -11.74) >consensus UCGCCAGAUGCUAUUGUAACUAUUAAU_UGUUGUUAUCCGCA_UCCUCUUACUAUAUUUAAUCAACAU___ACAUUUCAAUAGUCUCUGGGCAACCAU ...((((..(((((((.............................................................)))))))..))))........ ( -6.71 = -7.18 + 0.47)


| Location | 12,995,150 – 12,995,249 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.94 |
| Mean single sequence MFE | -22.00 |
| Consensus MFE | -14.04 |
| Energy contribution | -13.52 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12995150 99 + 20766785 UUCCGCA-UUCUCUUACUAUAUUUCAUCAAUAU---ACCUUUCAAUAGUCUCUGGGCAACCAUGAGUUUGAUCAGAAUGUUGAGGGUUUGGUGCCCUUCCUAA .((.(((-((((.....((((((.....)))))---)....((((....((((((....))).))).))))..))))))).))((((.....))))....... ( -24.80) >DroVir_CAF1 18214 97 + 1 -UCGUAAAUA-UAUUCCCAUAAUAACUACUU-U---GCUUUUUUGCAGUCACUUGGAAACCACGAGUUUGAUAAGAACGUUAAGGGUCUGAUACCAUUCCUCA -.......((-(...(((.((((........-(---((......)))((((((((.......)))))..)))......)))).)))....))).......... ( -13.20) >DroSec_CAF1 10518 99 + 1 AUCCGCA-UCCUUUUAUUAAGUUUAAUCAACAU---ACAUUUCAAUAGUCUCUGGGCAACCAUGAGUUUGACCAGAAUGUUGAGGGUUUGGUGCCCUUCCUCA .......-..................(((((((---.(...((((....((((((....))).))).))))...).)))))))((((.....))))....... ( -23.60) >DroSim_CAF1 9595 99 + 1 AUCCGCA-UCCUUUUAUUAUAUUUAAUCAACAU---ACACUUCAAUAGUCUCUGGGCAACCAUGAGUUUGACCAGAAUGUUGAGGGUUUGGUGCCCUUCCUCA .......-..................(((((((---.(...((((....((((((....))).))).))))...).)))))))((((.....))))....... ( -23.60) >DroEre_CAF1 9628 99 + 1 AUCCGCA-UUCGCUCACUAUAUUUUAUGAACCU---GCAUUUGAAUAGUCCCUGGGCAACCAUGAGUUCGACCAGAAUGUUGAGGGUUUGGUGCCCUUCCUCA .((.(((-((((((((((((.((..(((.....---.)))..)))))))...(((....))).)))).......)))))).))((((.....))))....... ( -23.51) >DroYak_CAF1 9772 102 + 1 AACCGCA-UCAUCUUACCAUAUUUCAUUAAUAUAUAACAUUUGAAUAGUCCCUGGGCAACCAUGAGUUCGACCAGAAUGUUGAGGGUUUGGUGCCCUUCCUCA ....(((-(((....((((((((.....))))).((((((((.....(..(((((....))).).)..).....))))))))..))).))))))......... ( -23.30) >consensus AUCCGCA_UCCUCUUACUAUAUUUAAUCAACAU___ACAUUUCAAUAGUCUCUGGGCAACCAUGAGUUUGACCAGAAUGUUGAGGGUUUGGUGCCCUUCCUCA .....................................................(((((.(((.(..((((((......))))))..).))))))))....... (-14.04 = -13.52 + -0.53)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:47 2006