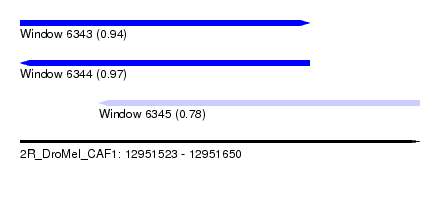
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,951,523 – 12,951,650 |
| Length | 127 |
| Max. P | 0.972674 |

| Location | 12,951,523 – 12,951,615 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 72.23 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -12.65 |
| Energy contribution | -12.85 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12951523 92 + 20766785 GC---------AAGUGCAAC-UGUAAGUGCAGCAGCGCUGCUUGAAAAACUGAGCGCCUGUUCCAAAGGCGCUGUAAAACUUAAAAACAACAAACAGACGC------G ((---------(..(((...-.)))..)))....((((((.(((........(((((((.......)))))))((...........))..))).))).)))------. ( -27.60) >DroVir_CAF1 3621 93 + 1 ---CAGCGA-GGCACACAGCAUGCAGUUGG-----CUGUGCUUUUUUCGCUGCGCGCCUGUUAAUAAGGCGCUGUAAAACUUAAAAACAACAAACAGACGC------G ---((((((-((((((((((.....)))).-----.))))))....)))))).((((((.......)))))).............................------. ( -29.50) >DroGri_CAF1 3247 97 + 1 ---CAGC-A-UGCAUGCAGC-UGUAGGGGA-----CUGAGCUUUUUUCGCUGUGCGCCUGUUAAUAAGGCGCUGUAAAACUUAAAAACAACAAACAGACGCGAGAGCG ---((((-.-((....))))-))(((....-----))).((((((..((((((((((((.......))))))(((...........)))....)))).)).)))))). ( -30.60) >DroEre_CAF1 3426 92 + 1 GC---------AAGUGCAGC-UGUAAGUGCAGCAGCGCUGCUUGAAAAACUGCGCGCCUGUUCCAAAGGCGCUGUAAAACUUAAAAACAACAAACAGACGC------G ((---------(.((((.((-(((....))))).)))))))..........((((((((.......))))))(((...........)))..........))------. ( -28.90) >DroMoj_CAF1 4742 93 + 1 GG-UAGCGA-GGC--ACAGCAUGCAGUUGG-----UUGUGCUGUUGGCGCUGUGCGCCUGUUAAUAAGGCGCUGUAAAACUUAAAAACAACAAACAGACGC------G ..-((((..-(((--(((((.........)-----)))))))))))(((((((((((((.......))))))(((...........)))....)))).)))------. ( -30.60) >DroAna_CAF1 3222 94 + 1 GCG-AGCAAGUAAAUGCAAC-UGUAAAUGU-----CGCUGCUUGGAAAACUGCGCGCCUGUUCCAAAGGCGCUGUAAAACUUAAAAACAACAAACAGACCC------- .((-((((.(..(.(((...-.)))....)-----..)))))))......(((((((((.......)))))).))).........................------- ( -20.30) >consensus GC__AGC_A__AAAUGCAGC_UGUAAGUGC_____CGCUGCUUGUAAAACUGCGCGCCUGUUAAAAAGGCGCUGUAAAACUUAAAAACAACAAACAGACGC______G ..............((((....(((.(((......))))))............((((((.......))))))))))................................ (-12.65 = -12.85 + 0.20)

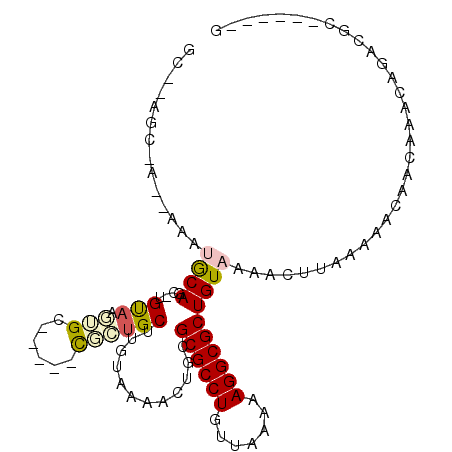

| Location | 12,951,523 – 12,951,615 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 72.23 |
| Mean single sequence MFE | -28.59 |
| Consensus MFE | -16.13 |
| Energy contribution | -15.97 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12951523 92 - 20766785 C------GCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUUGGAACAGGCGCUCAGUUUUUCAAGCAGCGCUGCUGCACUUACA-GUUGCACUU---------GC .------(((.((((..((................(((((((.......)))))))...........(((((...)))))))..)))-).)))....---------.. ( -27.30) >DroVir_CAF1 3621 93 - 1 C------GCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUAUUAACAGGCGCGCAGCGAAAAAAGCACAG-----CCAACUGCAUGCUGUGUGCC-UCGCUG--- (------(((((((((((((((...........)))))).......))))))))))((((((.....((((((-----(.........)))))))...-))))))--- ( -35.11) >DroGri_CAF1 3247 97 - 1 CGCUCUCGCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUAUUAACAGGCGCACAGCGAAAAAAGCUCAG-----UCCCCUACA-GCUGCAUGCA-U-GCUG--- ((((...(((((((((((((((...........)))))).......)))))))))..))))............-----.......((-((((....))-.-))))--- ( -25.81) >DroEre_CAF1 3426 92 - 1 C------GCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUUGGAACAGGCGCGCAGUUUUUCAAGCAGCGCUGCUGCACUUACA-GCUGCACUU---------GC .------(((((((((((((((...........))))).......))))))))))((((((......(((((...)))))......)-)))))....---------.. ( -31.51) >DroMoj_CAF1 4742 93 - 1 C------GCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUAUUAACAGGCGCACAGCGCCAACAGCACAA-----CCAACUGCAUGCUGU--GCC-UCGCUA-CC .------(((((((((((((((...........)))))).......)))))))))..((((...((((((((.-----.....))..))))))--...-.)))).-.. ( -28.41) >DroAna_CAF1 3222 94 - 1 -------GGGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUUGGAACAGGCGCGCAGUUUUCCAAGCAGCG-----ACAUUUACA-GUUGCAUUUACUUGCU-CGC -------((((.((((((.(((...........)))((((((.......)))))).........))))))(((-----((.......-)))))........)))-).. ( -23.40) >consensus C______GCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUAGGAACAGGCGCGCAGCGUUAAAAGCACAG_____CCACCUACA_GCUGCAUGC__U_GCU__GC .......(((((((((((((((...........)))))).......)))))))))(((((............................)))))............... (-16.13 = -15.97 + -0.16)

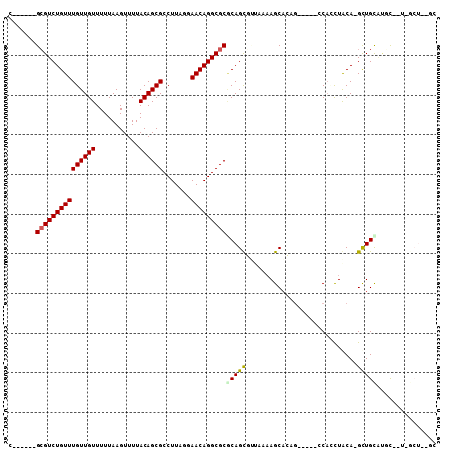
| Location | 12,951,548 – 12,951,650 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -18.02 |
| Energy contribution | -19.14 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12951548 102 - 20766785 CGC-AAACUAUUUUCCGCGCUGUAACUAAACGAGCGCGCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUUGGAACAGGCGCUCAGUUUUUCAAGCAGCG (((-............(((((((......)).)))))(((((((((((((((...........))))).......))))))))))...............))) ( -28.41) >DroVir_CAF1 3647 90 - 1 UGCAAAACUAUUUUG----CUUU---------CGCGCGCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUAUUAACAGGCGCGCAGCGAAAAAAGCACAG .(((((.....))))----)(((---------((((((((((((((((((((...........)))))).......))))))))))).))))))......... ( -32.51) >DroEre_CAF1 3451 102 - 1 CGC-AAACUAUUUUCCGCGCUGUAACUACACGAGCGCGCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUUGGAACAGGCGCGCAGUUUUUCAAGCAGCG ...-.............((((((........(((((((((((((((((((((...........))))).......)))))))))))).)))).....)))))) ( -30.73) >DroYak_CAF1 3329 102 - 1 CGC-AAACUAUUUUCCGCGCUGUAACUACACGAGCGAGCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUUGGAACAGGCGCUCAGUUUUUCAAGCAGCG ...-.............((((((........(((((((((((((((((((((...........))))).......)))))))))))).)))).....)))))) ( -29.43) >DroMoj_CAF1 4768 90 - 1 CUCAAAACUAUUUCG----CUUU---------CGCGCGCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUAUUAACAGGCGCACAGCGCCAACAGCACAA ..............(----((((---------.(((((((((((((((((((...........)))))).......)))))))))...)))).)).))).... ( -24.91) >DroAna_CAF1 3250 99 - 1 GGC-AAACUAUC-CGCGCGCUGUAACUAUUCC-GCG-GGGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUUGGAACAGGCGCGCAGUUUUCCAAGCAGCG (((-((((...(-(.((((..((....))..)-)))-.))...)))))))................((((((.......))))))((.(((.....))).)). ( -30.00) >consensus CGC_AAACUAUUUUCCGCGCUGUAACUA_ACGAGCGCGCGUCUGUUUGUUGUUUUUAAGUUUUACAGCGCCUUUGGAACAGGCGCGCAGUUUUUCAAGCAGCG .................((((((.........(((..(((((((((((((((...........)))))).......)))))))))...)))......)))))) (-18.02 = -19.14 + 1.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:35 2006