| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,938,740 – 12,938,849 |
| Length | 109 |
| Max. P | 0.716061 |

| Location | 12,938,740 – 12,938,849 |
|---|---|
| Length | 109 |
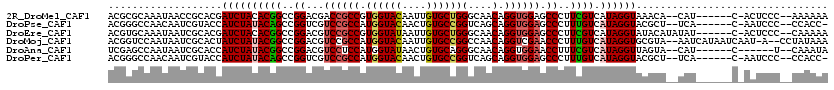
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.99 |
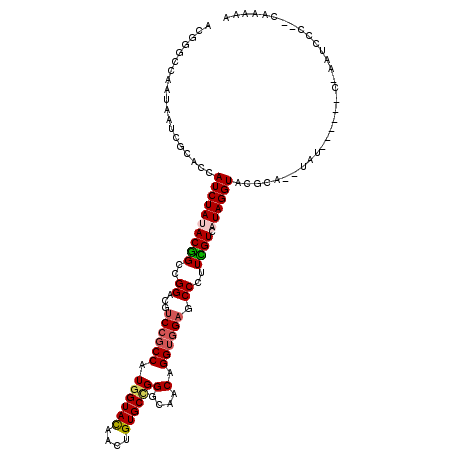
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -21.01 |
| Energy contribution | -21.15 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12938740 109 + 20766785 ACGCGCAAAUAACCGCACGAUCUACACGGCCGGACGACCGCCGUGGUACAAUUGUGCUGGGCAACAGGUGGAGCCCUUCGUCAUAGGUAAACA--CAU------C-ACUCCC--AAAAAA ...........((((((((((...((((((.((....)))))))).....))))))).((((..(....)..)))).........))).....--...------.-......--...... ( -31.90) >DroPse_CAF1 225 108 + 1 ACGGGCCAACAAUCGUACCAUCUAUACAGCCGGUCGUCCGCCAUGGUACAACUGUGCCGGUCAGCAGGUGGAGCCCUUUGUCAUAGGUACGCU--UCA------C-AAUCCC--CCACC- ..(((........((((((...(((((((..(((..((((((.((((((....))))))(....).)))))))))..)))).)))))))))..--...------.-...)))--.....- ( -32.43) >DroEre_CAF1 216 111 + 1 ACGUGCAAAUAAUCGCACGAUCUACACGGCCGGACGUCCGCCGUGGUAUAAUUGUGCUGGGCAACAGGUGGAGCCCUUCGUCAUAGGUAUACAUAUAU------C-ACUCCC--CAAAAA .(((((........))))).....((((((.((....))))))))(((((.((((((.((((..(....)..))))...).))))).)))))......------.-......--...... ( -36.30) >DroMoj_CAF1 153 115 + 1 ACGGUCCAAUAAUCGCACUAUCUAUACGGCCGGACGUCCGCCAUGGUACAAUUGUGCCGGCCAACAGGUCGAACCCUUUGUCAUAGGUGCGUA--AAUCAUAAUCAAU-A--CCUAUAAA ..(((........(((((...((((((((((((....)).....(((((....)))))))))...(((......)))..)).)))))))))..--.......)))...-.--........ ( -28.53) >DroAna_CAF1 201 104 + 1 UCGAGCCAAUAAUCGCACCAUCUAUACGGCCGGACGUCCUCCAUGGUAUAACUGUGCAGGGCAACAGGUGGAACCUUUCGUCAUAGGUUAGUA--CAU------C------U--CAAAUA ..(((............((((((....(......)(((((.(((((.....))))).)))))...))))))(((((........)))))....--...------)------)--)..... ( -25.50) >DroPer_CAF1 225 108 + 1 ACGGGCCAACAAUCGUACCAUCUAUACAGCCGGUCGUCCGCCAUGGUACAACUGUGCCGGUCAGCAGGUGGAGCCCUUUGUCAUAGGUACGCU--UCA------C-AAUCCC--CCACC- ..(((........((((((...(((((((..(((..((((((.((((((....))))))(....).)))))))))..)))).)))))))))..--...------.-...)))--.....- ( -32.43) >consensus ACGGGCCAAUAAUCGCACCAUCUAUACGGCCGGACGUCCGCCAUGGUACAACUGUGCCGGGCAACAGGUGGAGCCCUUCGUCAUAGGUACGCA__UAU______C_AAUCCC__CAAAAA ...................((((((((((..((...((((((.((((((....))))))(....).)))))).))..)))).))))))................................ (-21.01 = -21.15 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:30 2006