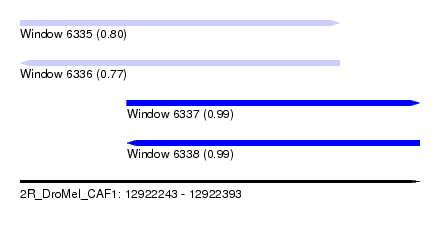
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,922,243 – 12,922,393 |
| Length | 150 |
| Max. P | 0.990044 |

| Location | 12,922,243 – 12,922,363 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -24.70 |
| Energy contribution | -26.08 |
| Covariance contribution | 1.37 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12922243 120 + 20766785 AGCGAAAUGAGUUACUCUGGUUUGGCCAAAAAUAAUUCUAUAACUGAGCUGCCAGAUCGCAUGCAGACUUAACAGUACACUGCUAACAGUUACUGUUAAAAGUACGCGCCAAUUUUGAAA ..((((((..((...((((((..(((((...(((....)))...)).)))))))))..((.(((....(((((((((.((((....)))))))))))))..))).))))..))))))... ( -29.70) >DroSec_CAF1 45564 120 + 1 AGCGAAAUGAGUUACCCUGGUUUGGGCAAAACUAAUUCUAUAACUGAGCUGCCAGAUCGCCGGCAGACUUAACAGUGCACUGCUAACAGUCACUGUUAAAAGUACGCGCCAAUUUUGGAA .(((....(((((((((......)))......))))))....(((...(((((........)))))..(((((((((.((((....))))))))))))).))).))).(((....))).. ( -35.70) >DroSim_CAF1 33790 120 + 1 AGCAAAAUGAGUUACCCUGGUUUGGCCAAAACUAAUUCUAUAACUGAGCUGCCAGAUCGCCGGCAGACUUAACAGUGCACUGCUAACAGUCACUGUUAAAAGUACGCGCCAAUUUUGAAA ..((((((..........((((((((........................))))))))...(((.((((((((((((.((((....))))))))))))..))).)..))).))))))... ( -32.36) >DroYak_CAF1 48109 119 + 1 AGCGAAAAGAGUUACUCUAGUUUUG-CACAAAUAUUUCUACAACUGAGCUGCCAGAUCGUCGGCAGAUUUAACAGUGGCCUGCUAACAGUCACUGUUAAAAGUACGCGGCAAUUUUGAAA .((((((..((.....))..)))))-).((((.(((.(..(.......(((((........))))).((((((((((((.((....)))))))))))))).....)..).)))))))... ( -33.10) >consensus AGCGAAAUGAGUUACCCUGGUUUGGCCAAAAAUAAUUCUAUAACUGAGCUGCCAGAUCGCCGGCAGACUUAACAGUGCACUGCUAACAGUCACUGUUAAAAGUACGCGCCAAUUUUGAAA .(((....((((((...(((.....)))....))))))....(((...(((((........)))))..(((((((((.((((....))))))))))))).))).)))............. (-24.70 = -26.08 + 1.37)

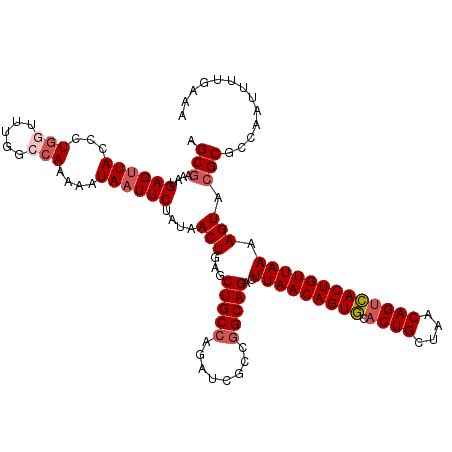
| Location | 12,922,243 – 12,922,363 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -25.99 |
| Energy contribution | -26.80 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12922243 120 - 20766785 UUUCAAAAUUGGCGCGUACUUUUAACAGUAACUGUUAGCAGUGUACUGUUAAGUCUGCAUGCGAUCUGGCAGCUCAGUUAUAGAAUUAUUUUUGGCCAAACCAGAGUAACUCAUUUCGCU ......((((((.(((((..((((((((((((((....)))).))))))))))..))).(((......)))))))))))...((.(((((((.((.....))))))))).))........ ( -31.90) >DroSec_CAF1 45564 120 - 1 UUCCAAAAUUGGCGCGUACUUUUAACAGUGACUGUUAGCAGUGCACUGUUAAGUCUGCCGGCGAUCUGGCAGCUCAGUUAUAGAAUUAGUUUUGCCCAAACCAGGGUAACUCAUUUCGCU ......((((((.((.....((((((((((((((....)))).))))))))))..((((((....))))))))))))))...(((...(..((((((......))))))..)..)))... ( -39.10) >DroSim_CAF1 33790 120 - 1 UUUCAAAAUUGGCGCGUACUUUUAACAGUGACUGUUAGCAGUGCACUGUUAAGUCUGCCGGCGAUCUGGCAGCUCAGUUAUAGAAUUAGUUUUGGCCAAACCAGGGUAACUCAUUUUGCU ......((((((.((.....((((((((((((((....)))).))))))))))..((((((....)))))))))))))).((((((.((((((((.....))))...)))).)))))).. ( -35.90) >DroYak_CAF1 48109 119 - 1 UUUCAAAAUUGCCGCGUACUUUUAACAGUGACUGUUAGCAGGCCACUGUUAAAUCUGCCGACGAUCUGGCAGCUCAGUUGUAGAAAUAUUUGUG-CAAAACUAGAGUAACUCUUUUCGCU .............(((....((((((((((.(((....)))..)))))))))).((((((......))))))((((((((((.((....)).))-))..))).)))..........))). ( -31.80) >consensus UUUCAAAAUUGGCGCGUACUUUUAACAGUGACUGUUAGCAGUGCACUGUUAAGUCUGCCGGCGAUCUGGCAGCUCAGUUAUAGAAUUAGUUUUGGCCAAACCAGAGUAACUCAUUUCGCU .............(((....((((((((((((((....)))).)))))))))).(((((((....)))))))..........((.(((.((((((.....))))))))).))....))). (-25.99 = -26.80 + 0.81)

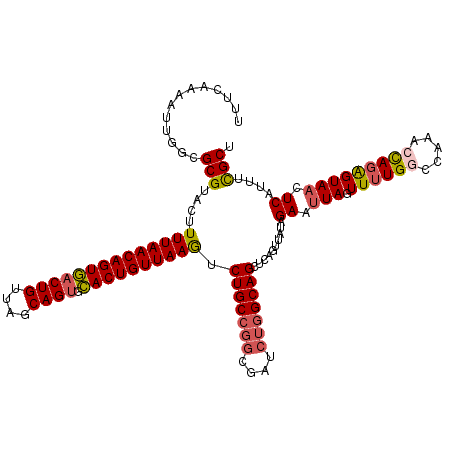
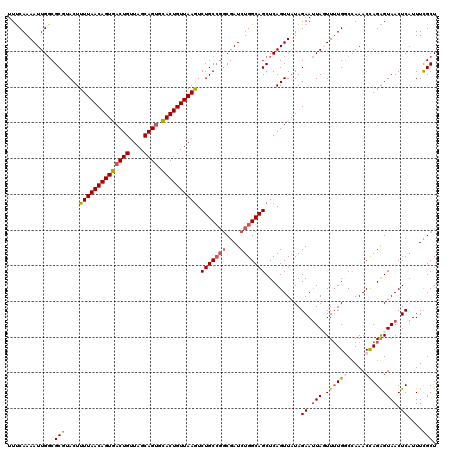
| Location | 12,922,283 – 12,922,393 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 90.39 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -27.83 |
| Energy contribution | -27.20 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12922283 110 + 20766785 UAACUGAGCUGCCAGAUCGCAUGCAGACUUAACAGUACACUGCUAACAGUUACUGUUAAAAGUACGCGCCAAUUUUGAAAGCUCGAAAGCCAAAGCUUACAAAAU-UAACU .....(((((..((((.(((.(((....(((((((((.((((....)))))))))))))..))).))).....))))..)))))(.((((....)))).).....-..... ( -30.10) >DroSec_CAF1 45604 111 + 1 UAACUGAGCUGCCAGAUCGCCGGCAGACUUAACAGUGCACUGCUAACAGUCACUGUUAAAAGUACGCGCCAAUUUUGGAAGCUCGAAAGCCAAAGCUUACAAAAUUUAACC .....(((((.(((((.....(((.((((((((((((.((((....))))))))))))..))).)..)))...))))).)))))(.((((....)))).)........... ( -35.90) >DroSim_CAF1 33830 111 + 1 UAACUGAGCUGCCAGAUCGCCGGCAGACUUAACAGUGCACUGCUAACAGUCACUGUUAAAAGUACGCGCCAAUUUUGAAAGCUCGAAUGCCAAAGCUUACCAAAUUUAACC ....((((((..((((.....(((.((((((((((((.((((....))))))))))))..))).)..)))...))))..)))))).......................... ( -30.30) >DroYak_CAF1 48148 111 + 1 CAACUGAGCUGCCAGAUCGUCGGCAGAUUUAACAGUGGCCUGCUAACAGUCACUGUUAAAAGUACGCGGCAAUUUUGAAAGCUCAAGAGUUAAAGCUUACAAAAUUGAAUU ...(((..(((((........))))).((((((((((((.((....))))))))))))))......)))((((((((.(((((..........))))).)))))))).... ( -35.70) >consensus UAACUGAGCUGCCAGAUCGCCGGCAGACUUAACAGUGCACUGCUAACAGUCACUGUUAAAAGUACGCGCCAAUUUUGAAAGCUCGAAAGCCAAAGCUUACAAAAUUUAACC ....((((((..((((......((..(((((((((((.((((....))))))))))))..)))..))......))))..)))))).((((....))))............. (-27.83 = -27.20 + -0.63)

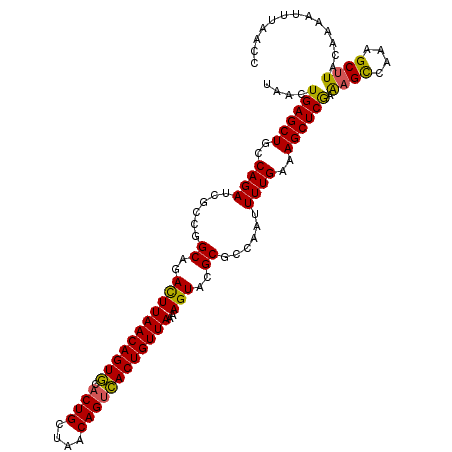
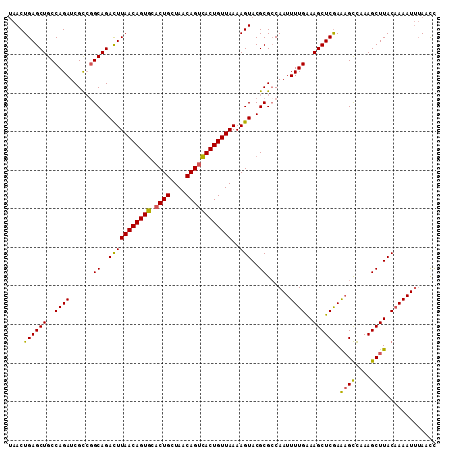
| Location | 12,922,283 – 12,922,393 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 90.39 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -32.03 |
| Energy contribution | -32.02 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.76 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12922283 110 - 20766785 AGUUA-AUUUUGUAAGCUUUGGCUUUCGAGCUUUCAAAAUUGGCGCGUACUUUUAACAGUAACUGUUAGCAGUGUACUGUUAAGUCUGCAUGCGAUCUGGCAGCUCAGUUA .((((-((((((.((((((........)))))).))))))))))(((((..((((((((((((((....)))).))))))))))..))).(((......)))))....... ( -38.20) >DroSec_CAF1 45604 111 - 1 GGUUAAAUUUUGUAAGCUUUGGCUUUCGAGCUUCCAAAAUUGGCGCGUACUUUUAACAGUGACUGUUAGCAGUGCACUGUUAAGUCUGCCGGCGAUCUGGCAGCUCAGUUA ...........(.((((....)))).)(((((.(((..((((.((.(((..((((((((((((((....)))).))))))))))..))))).)))).))).)))))..... ( -41.60) >DroSim_CAF1 33830 111 - 1 GGUUAAAUUUGGUAAGCUUUGGCAUUCGAGCUUUCAAAAUUGGCGCGUACUUUUAACAGUGACUGUUAGCAGUGCACUGUUAAGUCUGCCGGCGAUCUGGCAGCUCAGUUA .(((((.(((((.((((((........))))))))))).)))))........(((((((((((((....)))).)))))))))(.(((((((....))))))))....... ( -38.30) >DroYak_CAF1 48148 111 - 1 AAUUCAAUUUUGUAAGCUUUAACUCUUGAGCUUUCAAAAUUGCCGCGUACUUUUAACAGUGACUGUUAGCAGGCCACUGUUAAAUCUGCCGACGAUCUGGCAGCUCAGUUG ....((((((((.((((((........)))))).)))))))).........((((((((((.(((....)))..)))))))))).((((((......))))))........ ( -36.40) >consensus AGUUAAAUUUUGUAAGCUUUGGCUUUCGAGCUUUCAAAAUUGGCGCGUACUUUUAACAGUGACUGUUAGCAGUGCACUGUUAAGUCUGCCGGCGAUCUGGCAGCUCAGUUA ...........................(((((.(((..((((.((.(((..((((((((((((((....)))).))))))))))..))))).)))).))).)))))..... (-32.03 = -32.02 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:28 2006