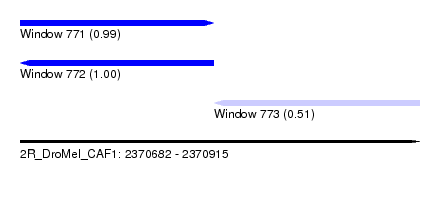
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,370,682 – 2,370,915 |
| Length | 233 |
| Max. P | 0.999812 |

| Location | 2,370,682 – 2,370,795 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -28.34 |
| Energy contribution | -28.38 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994873 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2370682 113 + 20766785 CAUGCAAAGG-------CGAAAUCACAAAAUACUGAAACGUGCUUUUUGACAUUUUUGCAAUGUAUUUAGGCACGUGCAAUAUCCAUCUCGGGGAGUGUGUGUGUGCGAUUCCUUUGCCA ...(((((((-------...((((........(((((((((((..............)).)))).)))))(((((..((...(((.......)))...))..)))))))))))))))).. ( -34.04) >DroSec_CAF1 20082 111 + 1 CAUGCAAAGG-------CGAAAUCACAAAACACUGAAACGUGCUUUUUGACAUUUUUGCAAUGUAUUUAGGCACGUGCAAUAUCCAUCCCGUUGAGUGU--GUGUGCGAUUCCUUUGCCA ...(((((((-------...((((.((..(((((...((((((((....(((((.....)))))....)))))))).((((.........)))))))))--...)).))))))))))).. ( -33.80) >DroSim_CAF1 20263 111 + 1 CAUGCAAAGG-------CGAAAUCACAAAACACUGAAACGUGCUUUUUGACAUUUUUGCAAUGUAUUUAGGCACGUGCAAUAUCCAUCCCGGGGAGUGU--GUGUGCGAUUCCUUUGCCA ...(((((((-------...((((.((..(((((...((((((((....(((((.....)))))....))))))))......(((......))))))))--...)).))))))))))).. ( -35.60) >DroEre_CAF1 19415 118 + 1 CAUGCAAAGGCGAAAGGCGAAAUCACAAAACACUGAAACGUGCUUUUUGACAUUUUUGCAAUGUAUUUAGGCACGUGCAACAUCCAUCUCAGCGAGUGU--GUGUGCGAUUCCUUGGCCA ........(((..((((...((((.((..(((((...((((((((....(((((.....)))))....))))))))((.............)).)))))--...)).)))))))).))). ( -33.82) >DroYak_CAF1 19543 111 + 1 CAUGCAAAGG-------CGAAAUCACAAAACACUGAAACGUGCUUUUUGACAUUUUUGCAAUGUAUUUAGGCACGUGCAAUAUCCAUCUCAGCGAGUGU--GUGUGCGAUUCCUUUGCCA ...(((((((-------...((((.((..(((((...((((((((....(((((.....)))))....))))))))((.............)).)))))--...)).))))))))))).. ( -34.02) >consensus CAUGCAAAGG_______CGAAAUCACAAAACACUGAAACGUGCUUUUUGACAUUUUUGCAAUGUAUUUAGGCACGUGCAAUAUCCAUCUCGGCGAGUGU__GUGUGCGAUUCCUUUGCCA ...(((((((.......((....((((..((((....((((((((....(((((.....)))))....))))))))..........((.....))))))...))))))...))))))).. (-28.34 = -28.38 + 0.04)

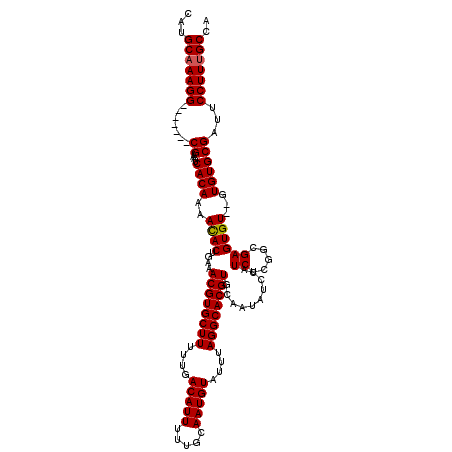
| Location | 2,370,682 – 2,370,795 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -33.04 |
| Consensus MFE | -30.50 |
| Energy contribution | -30.14 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2370682 113 - 20766785 UGGCAAAGGAAUCGCACACACACACUCCCCGAGAUGGAUAUUGCACGUGCCUAAAUACAUUGCAAAAAUGUCAAAAAGCACGUUUCAGUAUUUUGUGAUUUCG-------CCUUUGCAUG ..((((((((((((((......(((((...))).))(((((((.((((((......(((((.....)))))......))))))..))))))).)))))))...-------)))))))... ( -33.60) >DroSec_CAF1 20082 111 - 1 UGGCAAAGGAAUCGCACAC--ACACUCAACGGGAUGGAUAUUGCACGUGCCUAAAUACAUUGCAAAAAUGUCAAAAAGCACGUUUCAGUGUUUUGUGAUUUCG-------CCUUUGCAUG ..((((((((((((((...--.((..(....)..))(((((((.((((((......(((((.....)))))......))))))..))))))).)))))))...-------)))))))... ( -33.10) >DroSim_CAF1 20263 111 - 1 UGGCAAAGGAAUCGCACAC--ACACUCCCCGGGAUGGAUAUUGCACGUGCCUAAAUACAUUGCAAAAAUGUCAAAAAGCACGUUUCAGUGUUUUGUGAUUUCG-------CCUUUGCAUG ..((((((((((((((...--.((..(....)..))(((((((.((((((......(((((.....)))))......))))))..))))))).)))))))...-------)))))))... ( -34.60) >DroEre_CAF1 19415 118 - 1 UGGCCAAGGAAUCGCACAC--ACACUCGCUGAGAUGGAUGUUGCACGUGCCUAAAUACAUUGCAAAAAUGUCAAAAAGCACGUUUCAGUGUUUUGUGAUUUCGCCUUUCGCCUUUGCAUG .(((...(((((((((...--.....((((((((((....(((((.(((........))))))))...(((......)))))))))))))...))))))))))))....((....))... ( -30.30) >DroYak_CAF1 19543 111 - 1 UGGCAAAGGAAUCGCACAC--ACACUCGCUGAGAUGGAUAUUGCACGUGCCUAAAUACAUUGCAAAAAUGUCAAAAAGCACGUUUCAGUGUUUUGUGAUUUCG-------CCUUUGCAUG ..((((((((((((((...--.(((((...))).))(((((((.((((((......(((((.....)))))......))))))..))))))).)))))))...-------)))))))... ( -33.60) >consensus UGGCAAAGGAAUCGCACAC__ACACUCCCCGAGAUGGAUAUUGCACGUGCCUAAAUACAUUGCAAAAAUGUCAAAAAGCACGUUUCAGUGUUUUGUGAUUUCG_______CCUUUGCAUG ..((((((((((((((......(((((...))).))(((((((.((((((......(((((.....)))))......))))))..))))))).)))))))..........)))))))... (-30.50 = -30.14 + -0.36)

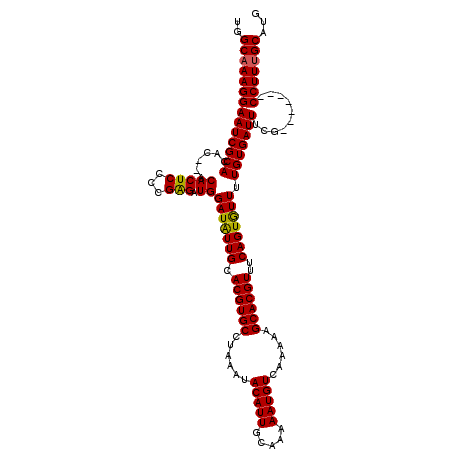
| Location | 2,370,795 – 2,370,915 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -23.30 |
| Energy contribution | -24.02 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2370795 120 - 20766785 UGGGUUUCGAUUUUGUUGACAAUUCACUUUGCAGCGGUAAUUGACCCUAAUGCUGUUGCUGCCGAUUCCGCUCGGCUUUCGCCUUUAAAUCGAUAUCACUUUUGGCAGCUAAAUUCAAUU .(((..((((((((((((.(((......))))))))).)))))))))..........((((((((........(((....)))........(....)....))))))))........... ( -30.90) >DroSec_CAF1 20193 120 - 1 AGGGUUUCGAUUUUGUUGACAAUUCACUUUGCAGCGGUAAUUGACCCUAAUGCUGUUGCUGCUGAUUCCGCUCGGUUUUCGCCUUUUAAUCGAUAUCACUUUUGGCAGCUAAAUUCAAUU ((((..((((((((((((.(((......))))))))).)))))))))).........(((((..(......((((((..........))))))........)..)))))........... ( -29.64) >DroSim_CAF1 20374 120 - 1 CGGGUUUCGAUUUUGUUGACAAUUCACUUUGCAGCGGUAAUUGACCCUAAUGCUGUUGCUGCUGAUUCCGCUCGGCUUUCGCCUUUAAAUCGAUAUCACUUUUGGCAGCUAAAUUCAAUU .(((..((((((((((((.(((......))))))))).)))))))))..........(((((..(........(((....)))........(....)....)..)))))........... ( -28.70) >DroEre_CAF1 19533 120 - 1 GGGGCUUCGAUUUUGUUGACAAUUCACUUUGCAGCGGUAAUUGACCCUAAUGCUGUUGCUCCUGAUUCCGCUCGGCUCCCGCCUUUAAAUCGAUAUCACUUUUGGCAGCUAAAUUCAAUU ((((..((((((((((((.(((......))))))))).))))))))))...((((((.....((((..((...(((....))).......))..)))).....))))))........... ( -28.50) >DroYak_CAF1 19654 109 - 1 UGGGUUUCGAUUUUGUUGACAAUUCACUUUGCAGCGGUAAUUGACCCUAAUGCUGUUGCUCCUGAUUCC-----------GCCUUUAAAUCGAUAUCACUUUUGGCAGCUAAAUUCAAUU .(((..((((((((((((.(((......))))))))).)))))))))....((((((.....((((..(-----------(.........))..)))).....))))))........... ( -23.50) >consensus UGGGUUUCGAUUUUGUUGACAAUUCACUUUGCAGCGGUAAUUGACCCUAAUGCUGUUGCUGCUGAUUCCGCUCGGCUUUCGCCUUUAAAUCGAUAUCACUUUUGGCAGCUAAAUUCAAUU .(((..((((((((((((.(((......))))))))).)))))))))..........((((((((........(((....)))........(....)....))))))))........... (-23.30 = -24.02 + 0.72)

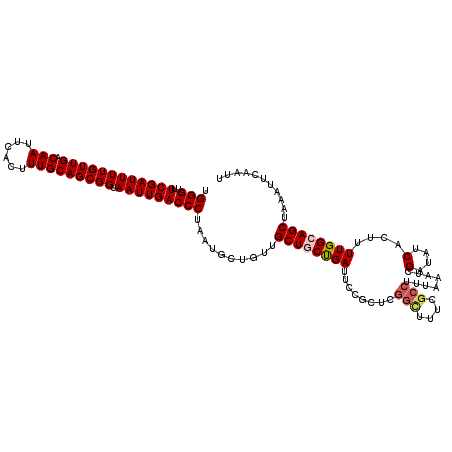
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:25 2006