
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,889,209 – 12,889,308 |
| Length | 99 |
| Max. P | 0.809095 |

| Location | 12,889,209 – 12,889,308 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.77 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -9.66 |
| Energy contribution | -10.13 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
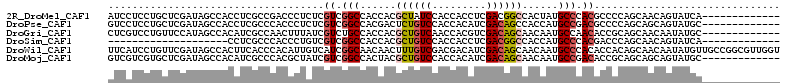
>2R_DroMel_CAF1 12889209 99 + 20766785 AUCCUCCUGCUCGAUAGCCACCUCGCCGACCCUCUCGUCGGCCACCACGCUAUCCACCACCUCGACGGCCACUAUGCCCACGCCCCAGCAACAGUAUCA------------- .......((((.((((((......((((((......))))))......))))))............(((......)))........)))).........------------- ( -24.40) >DroPse_CAF1 11993 99 + 1 GUCCUCCUGCUCGAUAGCCACCUCGCCCACCCUCUCGUCGGCCACGACUCUGUCCACCACAUCGACAGCCACCAUGCCGACGCCCCAGCAGCAGUAUGC------------- ...((.(((((.(...((......)).........(((((((.......(((((.........))))).......)))))))..).))))).)).....------------- ( -24.24) >DroGri_CAF1 28 99 + 1 CUCGUCCUGUUCCAUAGCCACAUCGCCAACUUUAUCGUCUGCCACCACGCUGUCAACCACGUCGACAGCAACAAUGCCAACACCGCAGCAACAAUAUGC------------- ............((((((......))..........((.(((......((((((.((...)).)))))).....(((.......))))))))..)))).------------- ( -15.00) >DroSim_CAF1 2 79 + 1 --------------------CCUCGCCCACCCUGUCGUCGGCCACCACGCUGUCCACCACCUCGACGGCCACCAUGCCCACGACCCAGCAACAGUAUCA------------- --------------------....((.......(((((.(((......((((((.........))))))......))).)))))...))..........------------- ( -19.00) >DroWil_CAF1 16296 112 + 1 UUCAUCCUGUUCGAUAGCCACUUCACCCACAUUGUCAUCGGCAACAACUUUGUCGACGACAUCGACAGCAACAAUGCCCACACCACAGCAACAAUAUGUUGCCGGCGUUGGU ......((((.((((.(......).........(((.(((((((.....))))))).)))))))))))...(((((((.........(((((.....))))).))))))).. ( -29.70) >DroMoj_CAF1 15728 99 + 1 GUCGUCGUGCUCGAUAGCCACAUCGCCCACGCUAUCGUCGGCCACUACGCUGUCCACCACAUCGACAGCAACAAUGCCGACACCGCAGCAGCAGUAUGC------------- .....((((((.((((((............))))))((((((......((((((.........))))))......))))))...((....)))))))).------------- ( -31.30) >consensus GUCCUCCUGCUCGAUAGCCACCUCGCCCACCCUAUCGUCGGCCACCACGCUGUCCACCACAUCGACAGCAACAAUGCCCACACCCCAGCAACAGUAUGC_____________ ....................................((.(((......((((((.........))))))......))).))............................... ( -9.66 = -10.13 + 0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:08 2006