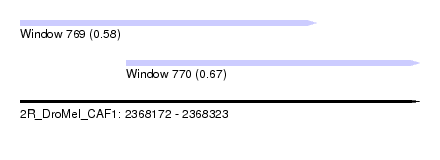
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,368,172 – 2,368,323 |
| Length | 151 |
| Max. P | 0.668892 |

| Location | 2,368,172 – 2,368,284 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -25.84 |
| Energy contribution | -28.32 |
| Covariance contribution | 2.48 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.581032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
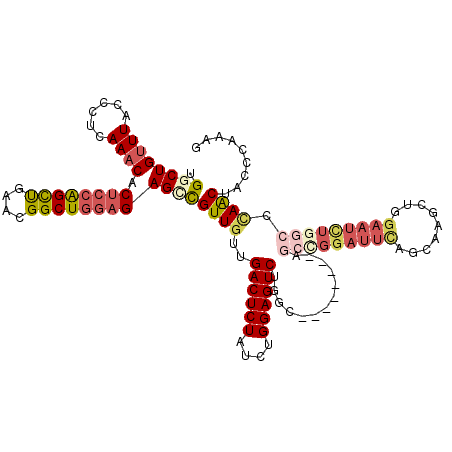
>2R_DroMel_CAF1 2368172 112 + 20766785 UGGCUGUUUACCCUCAAACACUCCAGCUGAACGGCUGGAGAGCUGUUGUUGACUCUAUCUGGAGUCUGGC--------AGGUGGAUUCAGCAAGCUGGAAUCUGGCACAACUACCCAAAG .((((((((......)))).((((((((....))))))))))))(((((.((((((....)))))).(.(--------((((...(((((....)))))))))).))))))......... ( -41.00) >DroSec_CAF1 17616 112 + 1 UGGCUGUUUACCCUCAAACACUCCAGCUGAACGGCUGGAGAGUCGUUGUUGACUCUAUCUGGAGUCUGGC--------AGCCGGAUUUAGCAAGCUGGAAUCUGGCCCAACUACCCAAAG .((((((((......)))).((((((((....))))))))))))((((..((((((....))))))....--------.(((((((((((....)).))))))))).))))......... ( -42.40) >DroSim_CAF1 17764 112 + 1 UGGCUGUUUACCCUCAAACACUCGAGCUGAACGGCUGGAGAGCCGUUGUUGACUCUAUCUGGAGUCUGGC--------AGCCGGAUUCAGCAAUCUGGAAUCUGGCCCAACUACCCAAAG .((((((((......)))).(((.((((....)))).)))))))((((..((((((....))))))....--------.(((((((((((....)).))))))))).))))......... ( -41.00) >DroEre_CAF1 17134 96 + 1 CGUCUGUUUACCAGCAAACACUCCGGCUGAAUGGCUG-CGAGCCGUUGUUGACUCUAUCUGGAGUCUGGC----------------------CGCUGGAAUCUGGCC-AGCUACCCAAAG .((.(((......))).)).....((((.((((((..-...))))))...((((((....)))))).)))----------------------)(((((.......))-)))......... ( -31.10) >DroYak_CAF1 16986 120 + 1 GGUCUGCUUACCCUCAAACACUCCAGUCGAAUGGCUGGAGAGCCGUUGUUGACUCUAUCUGGAGUCUGGGAGUCUGGCAUCGGGAUUCUGCAAGCUGGAAUUUGGCUUAACUCUGCAAAG ....(((...(((.......((((((((....)))))))).((((((.(.((((((....)))))).).))...))))...))).......((((..(...)..))))......)))... ( -37.40) >consensus UGGCUGUUUACCCUCAAACACUCCAGCUGAACGGCUGGAGAGCCGUUGUUGACUCUAUCUGGAGUCUGGC________AGCCGGAUUCAGCAAGCUGGAAUCUGGCCCAACUACCCAAAG .((((((((......)))).((((((((....))))))))))))((((..((((((....)))))).............(((((((((.........))))))))).))))......... (-25.84 = -28.32 + 2.48)

| Location | 2,368,212 – 2,368,323 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.29 |
| Mean single sequence MFE | -38.82 |
| Consensus MFE | -20.12 |
| Energy contribution | -23.48 |
| Covariance contribution | 3.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
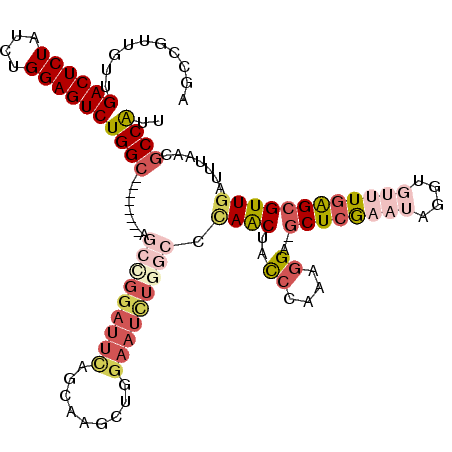
>2R_DroMel_CAF1 2368212 111 + 20766785 AGCUGUUGUUGACUCUAUCUGGAGUCUGGC--------AGGUGGAUUCAGCAAGCUGGAAUCUGGCACAACUACCCAAAGGA-GCUCGAAUAGGUGUUUGAGCGUUGAUUUAACGCCAUU ((((..((((((.((((((((........)--------))))))).))))))))))......((((.......((....)).-((((((((....))))))))...........)))).. ( -36.80) >DroSec_CAF1 17656 111 + 1 AGUCGUUGUUGACUCUAUCUGGAGUCUGGC--------AGCCGGAUUUAGCAAGCUGGAAUCUGGCCCAACUACCCAAAGGA-GCUCGAAUAGGUGUUUGAGCGUUGAUUUAACGCCAUU ..........((((((....))))))((((--------.(((((((((((....)).))))))))).((((..((....)).-((((((((....)))))))))))).......)))).. ( -40.20) >DroSim_CAF1 17804 111 + 1 AGCCGUUGUUGACUCUAUCUGGAGUCUGGC--------AGCCGGAUUCAGCAAUCUGGAAUCUGGCCCAACUACCCAAAGGA-GCUCGAAUAGGUGUUUGAGCGUUGAUUUAACGCCAUU ..........((((((....))))))((((--------.(((((((((((....)).))))))))).((((..((....)).-((((((((....)))))))))))).......)))).. ( -40.50) >DroEre_CAF1 17173 97 + 1 AGCCGUUGUUGACUCUAUCUGGAGUCUGGC----------------------CGCUGGAAUCUGGCC-AGCUACCCAAAGGAAGCUGGCCAAGGUGUUUGGGCGUUUAUUUAACGCCGUU .(((((.(((((((((....)))))).)))----------------------.)).......(((((-((((.((....)).))))))))).))).....((((((.....))))))... ( -42.10) >DroYak_CAF1 17026 120 + 1 AGCCGUUGUUGACUCUAUCUGGAGUCUGGGAGUCUGGCAUCGGGAUUCUGCAAGCUGGAAUUUGGCUUAACUCUGCAAAGGAACCUCACCACGGUGUUUGAGCGUUGAUUUGACGCCAUU .(((((.((.((((((....))))))..(((((..(((....(((((((.......))))))).)))..))))).............)).)))))...((.(((((.....))))))).. ( -34.50) >consensus AGCCGUUGUUGACUCUAUCUGGAGUCUGGC________AGCCGGAUUCAGCAAGCUGGAAUCUGGCCCAACUACCCAAAGGA_GCUCGAAUAGGUGUUUGAGCGUUGAUUUAACGCCAUU ..........((((((....))))))((((.........(((((((((.........))))))))).((((..((....))..((((((((....)))))))))))).......)))).. (-20.12 = -23.48 + 3.36)

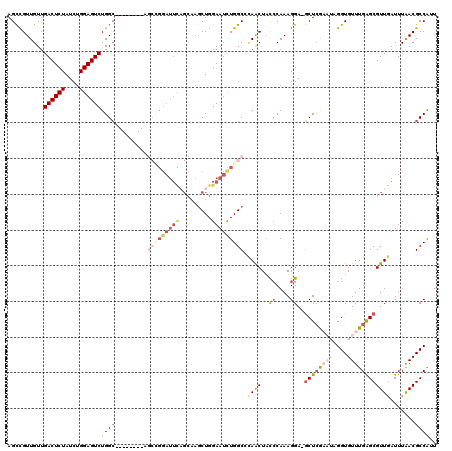
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:22 2006