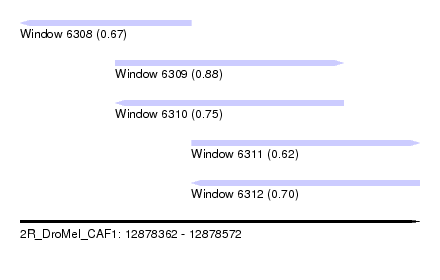
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,878,362 – 12,878,572 |
| Length | 210 |
| Max. P | 0.881214 |

| Location | 12,878,362 – 12,878,452 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.16 |
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -17.32 |
| Energy contribution | -18.83 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12878362 90 - 20766785 UGAGCCUGCAUUUGCAGUAGUGUUGCCAUGGAAACGGCUUGA---UGCUGGGCGGGCUGAACAGGCUG------------------UGCACCAGCUGCUGCGGCCACAUAG (.(((((((.....(((((.....(((..(....))))....---))))).))))))).)...(((((------------------((((.....))).))))))...... ( -35.40) >DroPse_CAF1 1542 90 - 1 UGAGCCUGCAUCUGCAGCAGGGUGGCCAUGGAGACGGCCUGC---GGCUGUGGCGCCUGGACGGGCUG------------------CGGGGCAGCUGCAGCGGCCACGAAU ....(((((.......)))))((((((.((....))((.(((---((((((.((((((....)))).)------------------)...))))))))))))))))).... ( -48.70) >DroSec_CAF1 1506 90 - 1 UGAGCCUGCAUUUGCAGUAGUGUUGCCAUGGAAACGGCUUGA---UGAUGGGCGGCCUGAACAGGCUG------------------UGCACCAGCUGCUGCGGCCACAUAG ((.(((.......((((((((...(((..(....))))....---((.((.(((((((....))))))------------------).)).)))))))))))))))..... ( -35.31) >DroEre_CAF1 1593 90 - 1 UGCGCCUGCAUUUGCAGUAAUGUGGCCAUGGAAACGGCCUGA---GGCUGGGCGGCCUGAACCGGCUG------------------CGCACCAGCUGCUGCGGCCACAUAG .....((((....))))..((((((((.((....))((....---((((((((((((......)))))------------------)...))))))...)))))))))).. ( -44.10) >DroWil_CAF1 1680 107 - 1 -GGCUCUGCAUUUGCAAGAGGGCAGCCAUUGACAUGGUCUGUCCGGGCAUGGGAGGUUGCACGGGUUGCGGCACUGGGGCUGGCGGUGGGGCAGCUGGAUUGGC---AAAG -.(((((((....)))...((((((((((....)))).))))))))))........((((.(.((((((..(((((.......)))))..)))))).)....))---)).. ( -40.60) >DroYak_CAF1 1602 90 - 1 UGUGCCUGCAUUUGCAUCAAUGUGGCCAUGGAAACGGCUUGA---GGCUGGGCGGCCUGAACGGGCUG------------------GGCACCAGCUGCUGCGGCCACAUAG ......(((....)))...((((((((.((....))((....---(((((((((((((....))))).------------------.)).))))))...)))))))))).. ( -41.70) >consensus UGAGCCUGCAUUUGCAGUAGUGUGGCCAUGGAAACGGCCUGA___GGCUGGGCGGCCUGAACGGGCUG__________________UGCACCAGCUGCUGCGGCCACAUAG ...((.(((....))))).((((.(((.......(((((......))))).(((((.......(((((.......................))))))))))))).)))).. (-17.32 = -18.83 + 1.50)
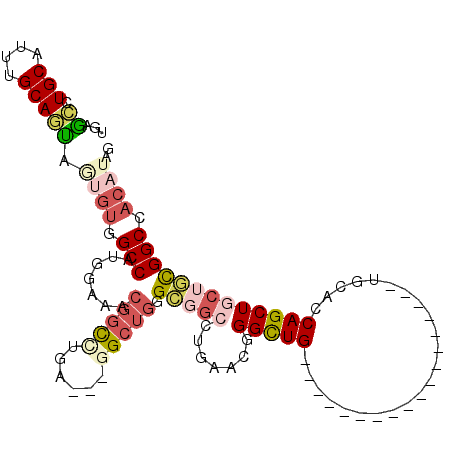
| Location | 12,878,412 – 12,878,532 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -42.62 |
| Consensus MFE | -26.78 |
| Energy contribution | -27.59 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12878412 120 + 20766785 AAGCCGUUUCCAUGGCAACACUACUGCAAAUGCAGGCUCAGGCCCAGGCGCAGGCAGCACAAGUUCAGGCCCAAGCUCAGGCUCAAGCUCAAGCUCAAGCUCAGGCCCAGGCUCAGGCCC ..(((((....))))).......((((...(((.(((....)))...)))...))))..........((((..((((..((((..((((........))))..))))..))))..)))). ( -43.60) >DroSec_CAF1 1556 120 + 1 AAGCCGUUUCCAUGGCAACACUACUGCAAAUGCAGGCUCAGGCCCAGGCGCAGGCGGCACAAGUCCAGGCCCAAGCUCAGGCUCAAGCUCAAGCUCAGGCCCAGGCCCAGGCUCAGGCCC ..(((((....))))).......((((....)))).....((((..(((...((((((....)))..((((..((((..(((....)))..))))..))))...)))...)))..)))). ( -46.10) >DroEre_CAF1 1643 105 + 1 AGGCCGUUUCCAUGGCCACAUUACUGCAAAUGCAGGCGCAGGCACAGGCCCAGGCG---------CAGGCACAAGUUCAGGCUCAGGCCCAGGCUCAA------GCCCAGGCUCAGGCCC .((((((....))))))......((((....))))((((.(((....)))...)))---------).(((...((((..((((..(((....)))..)------)))..))))...))). ( -46.00) >DroYak_CAF1 1652 99 + 1 AAGCCGUUUCCAUGGCCACAUUGAUGCAAAUGCAGGCACAGGCACAAGUCCAGG---------------CCCAAGCUCAGGCUCAGGCUCAAGCUCAA------GCCCAGGCUCAGGCCC ..(((...((.(((....))).))(((....))))))...(((....)))..((---------------((..((((..((((..(((....)))..)------)))..))))..)))). ( -34.80) >consensus AAGCCGUUUCCAUGGCAACACUACUGCAAAUGCAGGCUCAGGCACAGGCCCAGGCG_________CAGGCCCAAGCUCAGGCUCAAGCUCAAGCUCAA______GCCCAGGCUCAGGCCC ..(((((....)))))........(((....)))((((..((((..(((...(((.............)))..((((..(((....)))..)))).........)))..))))..)))). (-26.78 = -27.59 + 0.81)

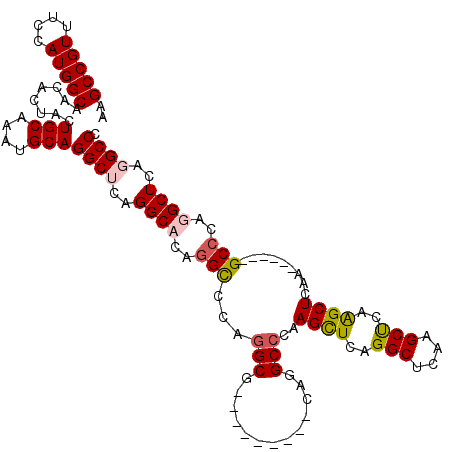
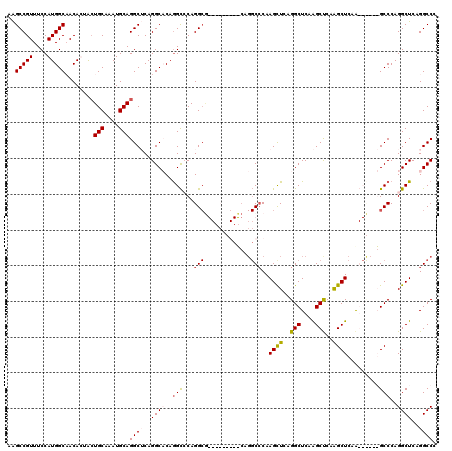
| Location | 12,878,412 – 12,878,532 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.57 |
| Mean single sequence MFE | -50.90 |
| Consensus MFE | -32.22 |
| Energy contribution | -34.91 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12878412 120 - 20766785 GGGCCUGAGCCUGGGCCUGAGCUUGAGCUUGAGCUUGAGCCUGAGCUUGGGCCUGAACUUGUGCUGCCUGCGCCUGGGCCUGAGCCUGCAUUUGCAGUAGUGUUGCCAUGGAAACGGCUU (((((..(((..((((..((((((......))))))..))))..)))..))))).(((...((((((.(((....((((....)))))))...))))))..)))(((..(....)))).. ( -54.40) >DroSec_CAF1 1556 120 - 1 GGGCCUGAGCCUGGGCCUGGGCCUGAGCUUGAGCUUGAGCCUGAGCUUGGGCCUGGACUUGUGCCGCCUGCGCCUGGGCCUGAGCCUGCAUUUGCAGUAGUGUUGCCAUGGAAACGGCUU ((((((..((.(((((..(((((..(((((..((....))..)))))..)))))((.(....)))))))).))..))))))((((((((....))).............(....)))))) ( -56.60) >DroEre_CAF1 1643 105 - 1 GGGCCUGAGCCUGGGC------UUGAGCCUGGGCCUGAGCCUGAACUUGUGCCUG---------CGCCUGGGCCUGUGCCUGCGCCUGCAUUUGCAGUAAUGUGGCCAUGGAAACGGCCU (((((.(.(((..(((------....)))..)))).).))))......((((..(---------(((..((((....))))))))..))))..(((....)))((((..(....))))). ( -46.70) >DroYak_CAF1 1652 99 - 1 GGGCCUGAGCCUGGGC------UUGAGCUUGAGCCUGAGCCUGAGCUUGGG---------------CCUGGACUUGUGCCUGUGCCUGCAUUUGCAUCAAUGUGGCCAUGGAAACGGCUU (((((..(((..((((------((..((....))..))))))..)))..))---------------)))........(((((.(((.(((((......)))))))))).(....)))).. ( -45.90) >consensus GGGCCUGAGCCUGGGC______UUGAGCUUGAGCCUGAGCCUGAGCUUGGGCCUG_________CGCCUGCGCCUGGGCCUGAGCCUGCAUUUGCAGUAAUGUGGCCAUGGAAACGGCUU ((((((..((((((((....((((((((((..((....))..)))))))))).............))))))))..))))))..(((.(((((......)))))......(....)))).. (-32.22 = -34.91 + 2.69)

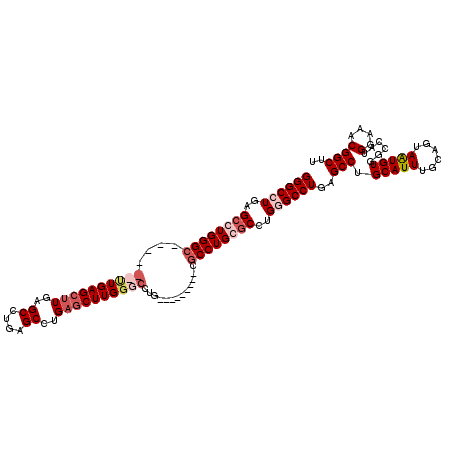
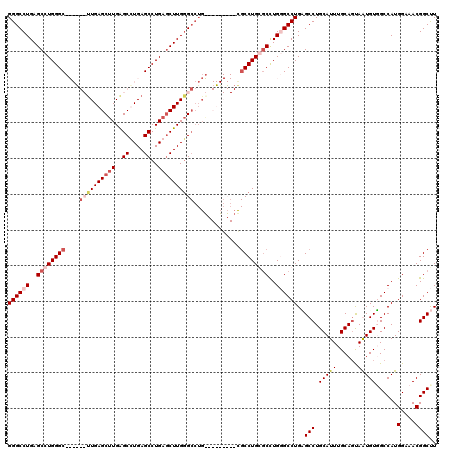
| Location | 12,878,452 – 12,878,572 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -45.22 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.10 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12878452 120 + 20766785 GGCCCAGGCGCAGGCAGCACAAGUUCAGGCCCAAGCUCAGGCUCAAGCUCAAGCUCAAGCUCAGGCCCAGGCUCAGGCCCAAAUGCGCCAGUUGGGUUCCCUGCCCAGUUCAGGACUAGC ((.((.((((((.(.(((....)))).((((..((((..((((..((((........))))..))))..))))..))))....))))))..((((((.....))))))....)).))... ( -46.30) >DroSec_CAF1 1596 120 + 1 GGCCCAGGCGCAGGCGGCACAAGUCCAGGCCCAAGCUCAGGCUCAAGCUCAAGCUCAGGCCCAGGCCCAGGCUCAGGCCCAAAUGCGCCAGUUGGGUUCCCUGCCCAGUUCAGGUCUAGC ((((..((((((((((((....)))..((((..((((..(((....)))..))))..))))...)))..(((....)))....))))))..((((((.....))))))....)))).... ( -51.20) >DroEre_CAF1 1683 105 + 1 GGCACAGGCCCAGGCG---------CAGGCACAAGUUCAGGCUCAGGCCCAGGCUCAA------GCCCAGGCUCAGGCCCAAAUGCGCCAGUUGGGAUCCCUGCCCAGUUCAGGACUAAC ((((..((((((((((---------(((((...((((..((((..(((....)))..)------)))..))))...)))....))))))...)))).))..))))............... ( -42.10) >DroYak_CAF1 1692 99 + 1 GGCACAAGUCCAGG---------------CCCAAGCUCAGGCUCAGGCUCAAGCUCAA------GCCCAGGCUCAGGCCCAAAUGCGUCAGUUGGGCUCCCUGCCCAGUUCAGGACUAAC ......(((((.((---------------((..((((..((((..(((....)))..)------)))..))))..))))............((((((.....))))))....)))))... ( -41.30) >consensus GGCACAGGCCCAGGCG_________CAGGCCCAAGCUCAGGCUCAAGCUCAAGCUCAA______GCCCAGGCUCAGGCCCAAAUGCGCCAGUUGGGUUCCCUGCCCAGUUCAGGACUAAC (((.(((((...(((.............)))...)))..((((..((((...(((........)))...))))..))))....)).)))..((((((.....))))))............ (-27.34 = -27.10 + -0.25)

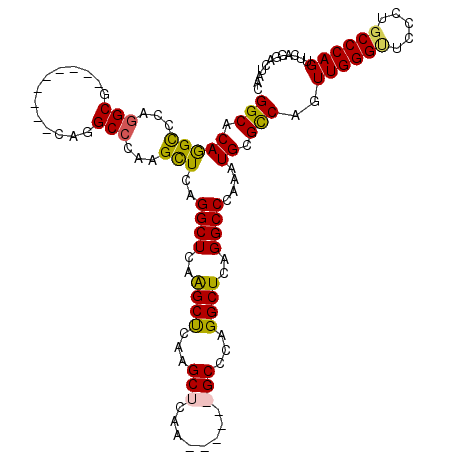
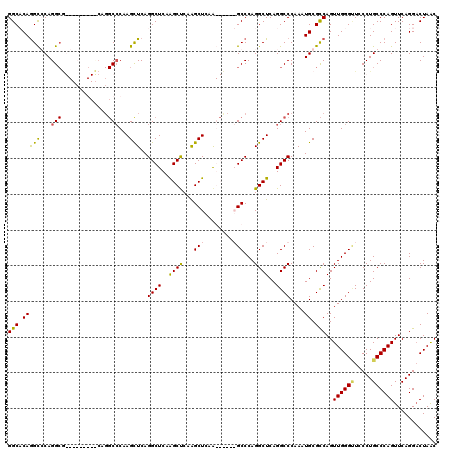
| Location | 12,878,452 – 12,878,572 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -53.23 |
| Consensus MFE | -31.73 |
| Energy contribution | -34.48 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12878452 120 - 20766785 GCUAGUCCUGAACUGGGCAGGGAACCCAACUGGCGCAUUUGGGCCUGAGCCUGGGCCUGAGCUUGAGCUUGAGCUUGAGCCUGAGCUUGGGCCUGAACUUGUGCUGCCUGCGCCUGGGCC ....((((......((((.((....))....((((((((..((((..(((..((((..((((((......))))))..))))..)))..))))..))..))))))))))......)))). ( -59.00) >DroSec_CAF1 1596 120 - 1 GCUAGACCUGAACUGGGCAGGGAACCCAACUGGCGCAUUUGGGCCUGAGCCUGGGCCUGGGCCUGAGCUUGAGCUUGAGCCUGAGCUUGGGCCUGGACUUGUGCCGCCUGCGCCUGGGCC ............(..(((.((....))....((((((((..((((..(((..((((..(((((.......).))))..))))..)))..))))..))..))))))......)))..)... ( -56.60) >DroEre_CAF1 1683 105 - 1 GUUAGUCCUGAACUGGGCAGGGAUCCCAACUGGCGCAUUUGGGCCUGAGCCUGGGC------UUGAGCCUGGGCCUGAGCCUGAACUUGUGCCUG---------CGCCUGGGCCUGUGCC ....(..(.(..(..(((.((....))....((((((((..((((.(.(((..(((------....)))..)))).).)))..))..))))))..---------.)))..)..).)..). ( -47.60) >DroYak_CAF1 1692 99 - 1 GUUAGUCCUGAACUGGGCAGGGAGCCCAACUGACGCAUUUGGGCCUGAGCCUGGGC------UUGAGCUUGAGCCUGAGCCUGAGCUUGGG---------------CCUGGACUUGUGCC ((((((.......(((((.....)))))))))))(((((..((((..(((..((((------((..((....))..))))))..)))..))---------------))..)....)))). ( -49.71) >consensus GCUAGUCCUGAACUGGGCAGGGAACCCAACUGGCGCAUUUGGGCCUGAGCCUGGGC______UUGAGCUUGAGCCUGAGCCUGAGCUUGGGCCUG_________CGCCUGCGCCUGGGCC ((((((((......)))).((....))...)))).......(((((..((((((((....((((((((((..((....))..)))))))))).............))))))))..))))) (-31.73 = -34.48 + 2.75)

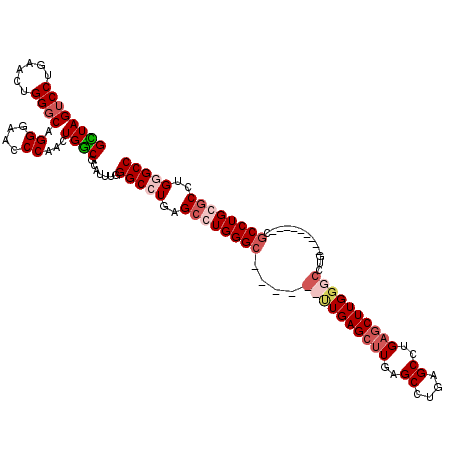
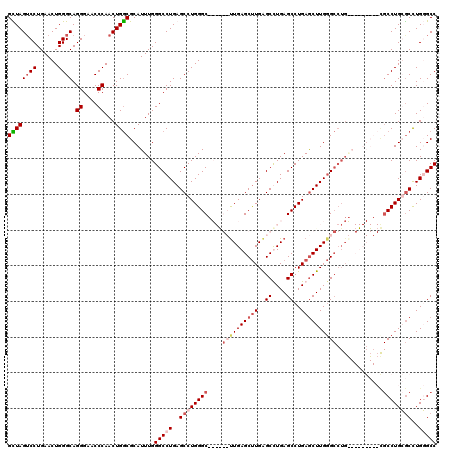
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:07:03 2006