| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,870,646 – 12,870,762 |
| Length | 116 |
| Max. P | 0.530966 |

| Location | 12,870,646 – 12,870,762 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
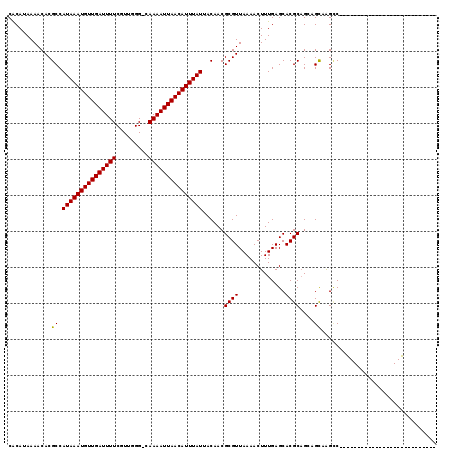
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -24.51 |
| Consensus MFE | -14.50 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.59 |
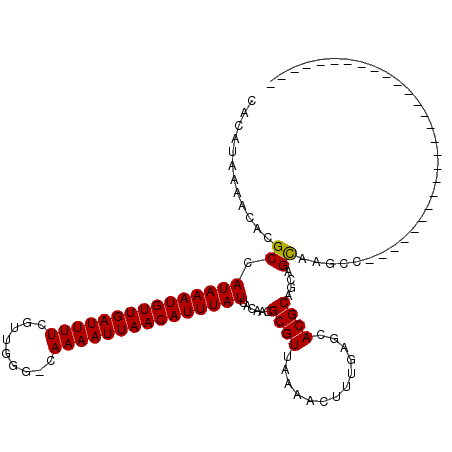
| Structure conservation index | 0.59 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
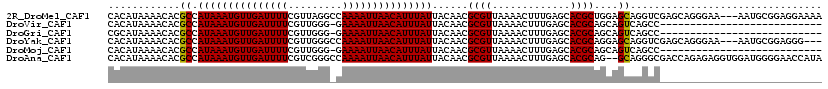
>2R_DroMel_CAF1 12870646 116 + 20766785 CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUUAGGCCAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAGCACGCUGGAGCAGGUCGAGCAGGGAA---AAUGCGGAGGAAAA .............(((((((((((((((((.(.....).))))))))))))))).....((((((....(((((..(((.(((...))))).)..)))))..---))))))..)).... ( -26.60) >DroVir_CAF1 5252 91 + 1 CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUUGGG-GAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAGCACGCAGCAGUCAGCC--------------------------- ............((.((((((((((((((((......-))))))))))))))))......((((.............)))).))........--------------------------- ( -20.22) >DroGri_CAF1 5160 91 + 1 CGCAUAAAACACGCCAUAAAUGUUGAUUUUCGUUGGG-GAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAGCACGCAGCAGUCAGCC--------------------------- .((.........(..((((((((((((((((......-))))))))))))))))..)...((((.............)))).))........--------------------------- ( -21.32) >DroYak_CAF1 3745 113 + 1 CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUUGGGCCAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAGCACGCAGGAGCAGGUCGAGCAGGGAA---AAUGCGGAGGG--- .............(((((((((((((((((.(.....).))))))))))))))).....((((((....(((((.((..((....))..))....)))))..---))))))..)).--- ( -25.30) >DroMoj_CAF1 5754 91 + 1 CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUUGGG-GAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAGCACGCAGCAGUCAGCC--------------------------- ............((.((((((((((((((((......-))))))))))))))))......((((.............)))).))........--------------------------- ( -20.22) >DroAna_CAF1 4718 117 + 1 CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUCGGGCCAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAGCACGCAG--GCAGGGCGACCAGAGAGGUGGAUGGGGAACCAUA .........(((((((((((((((((((((.(.....).)))))))))))))))......((((.............)))).)--)).((....))......))).((((....)))). ( -33.42) >consensus CACAUAAAACACGCCAUAAAUGUUGAUUUUCGUUGGG_CAAAAUUAACAUUUAUUACAACGCGUUAAAACUUUGAGCACGCAGCAGCAAGCC___________________________ ............((.(((((((((((((((.........)))))))))))))))......((((.............))))....))................................ (-14.50 = -14.25 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:53 2006