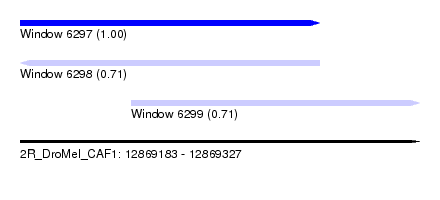
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,869,183 – 12,869,327 |
| Length | 144 |
| Max. P | 0.998907 |

| Location | 12,869,183 – 12,869,291 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.93 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -27.97 |
| Energy contribution | -28.47 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
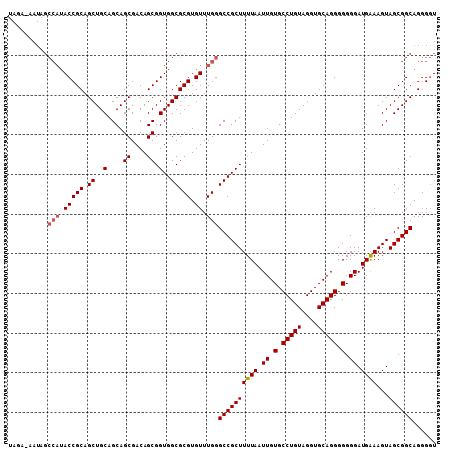
>2R_DroMel_CAF1 12869183 108 + 20766785 ACCCCUGCCGCUACUUCCAUCCCCCCCUGCACCUACAGGCACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUAUACUA ......((((((.............((((......))))..........))))))..........((((((((.((.((....)).)).)))))..)))......... ( -30.00) >DroVir_CAF1 3519 107 + 1 ACCCCUGCCGCUACUUUCAUCCCCCCCUGCACCUACAGGCACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUU-UCUA ......((((((.............((((......))))..........))))))..........((((((((.((.((....)).)).)))))..)))....-.... ( -30.00) >DroPse_CAF1 1737 94 + 1 AACCCUGCCGCUACUUUCAUCCCCCCCUGCACCUACAGGCACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGU-------------- ....((((.(((.............((((......))))...........(((((....(((.(((....))))))....)))))))).)))).-------------- ( -25.40) >DroGri_CAF1 3531 107 + 1 ACCCCUGCCGCUACUUUCAUCCCCCCCUGCACCUACAGGCACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUU-UCUA ......((((((.............((((......))))..........))))))..........((((((((.((.((....)).)).)))))..)))....-.... ( -30.00) >DroMoj_CAF1 4134 107 + 1 ACCCCUGCCGCUACUUUCAUCCCCCCCUGCACCUACAGGCACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUU-UCUA ......((((((.............((((......))))..........))))))..........((((((((.((.((....)).)).)))))..)))....-.... ( -30.00) >DroAna_CAF1 3152 108 + 1 ACCCCUGCCGCUACUUCCAUCCCCCCCUGCACCUACAGGCACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUAUACUA ......((((((.............((((......))))..........))))))..........((((((((.((.((....)).)).)))))..)))......... ( -30.00) >consensus ACCCCUGCCGCUACUUUCAUCCCCCCCUGCACCUACAGGCACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUU_UCUA ......((((((.............((((......))))..........))))))..........((((((((.((.((....)).)).)))))..)))......... (-27.97 = -28.47 + 0.50)



| Location | 12,869,183 – 12,869,291 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 93.93 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -36.92 |
| Energy contribution | -37.20 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12869183 108 - 20766785 UAGUAUAUAGCCAUACCGCAGCUGCAGCAGCGACAGCGGUGGCGCGUGUUUGGGCCGCUUUUAAUUGUGCCUGUAGGUGCAGGGGGGGAUGGAAGUAGCGGCAGGGGU ..........(((.(((((.((..(....((....)).)..))))).)).)))((((((((((.((.(.(((((....))))).).)).))))...))))))...... ( -38.20) >DroVir_CAF1 3519 107 - 1 UAGA-AAUAGCCAUACCGCAGCUGCAGCAGCGACAGCGGUGGCGCGUGUUUGGGCCGCUUUUAAUUGUGCCUGUAGGUGCAGGGGGGGAUGAAAGUAGCGGCAGGGGU ....-.....(((.(((((.((..(....((....)).)..))))).)).)))((((((((((.((.(.(((((....))))).).)).))))...))))))...... ( -38.60) >DroPse_CAF1 1737 94 - 1 --------------ACCGCAGCUGCAGCAGCGACAGCGGUGGCGCGUGUUUGGGCCGCUUUUAAUUGUGCCUGUAGGUGCAGGGGGGGAUGAAAGUAGCGGCAGGGUU --------------.((...(((((.((.((....)).))(((.((....)).)))(((((((.((.(.(((((....))))).).)).))))))).))))).))... ( -34.60) >DroGri_CAF1 3531 107 - 1 UAGA-AAUAGCCAUACCGCAGCUGCAGCAGCGACAGCGGUGGCGCGUGUUUGGGCCGCUUUUAAUUGUGCCUGUAGGUGCAGGGGGGGAUGAAAGUAGCGGCAGGGGU ....-.....(((.(((((.((..(....((....)).)..))))).)).)))((((((((((.((.(.(((((....))))).).)).))))...))))))...... ( -38.60) >DroMoj_CAF1 4134 107 - 1 UAGA-AAUAGCCAUACCGCAGCUGCAGCAGCGACAGCGGUGGCGCGUGUUUGGGCCGCUUUUAAUUGUGCCUGUAGGUGCAGGGGGGGAUGAAAGUAGCGGCAGGGGU ....-.....(((.(((((.((..(....((....)).)..))))).)).)))((((((((((.((.(.(((((....))))).).)).))))...))))))...... ( -38.60) >DroAna_CAF1 3152 108 - 1 UAGUAUAUAGCCAUACCGCAGCUGCAGCAGCGACAGCGGUGGCGCGUGUUUGGGCCGCUUUUAAUUGUGCCUGUAGGUGCAGGGGGGGAUGGAAGUAGCGGCAGGGGU ..........(((.(((((.((..(....((....)).)..))))).)).)))((((((((((.((.(.(((((....))))).).)).))))...))))))...... ( -38.20) >consensus UAGA_AAUAGCCAUACCGCAGCUGCAGCAGCGACAGCGGUGGCGCGUGUUUGGGCCGCUUUUAAUUGUGCCUGUAGGUGCAGGGGGGGAUGAAAGUAGCGGCAGGGGU ..........(((.(((((.((..(....((....)).)..))))).)).)))((((((((((.((.(.(((((....))))).).)).))))...))))))...... (-36.92 = -37.20 + 0.28)

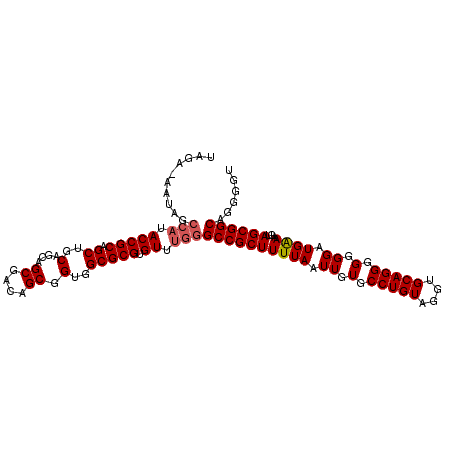
| Location | 12,869,223 – 12,869,327 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.08 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12869223 104 + 20766785 ACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUAUACUAUAUAUUUUU-CUAUACUUUUCGAACCUUCUCUUU--GCG ..........(((.........)))((((((((.((.((....)).)).)))))..)))..................-........................--... ( -20.70) >DroVir_CAF1 3559 93 + 1 ACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUU-UCUAUAUUUACUU-UUAUACUU---------UCUCUUU--C-U .....((((((((.........)))((((((((.((.((....)).)).)))))..)))....-...........))-))).....---------.......--.-. ( -20.80) >DroGri_CAF1 3571 93 + 1 ACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUU-UCUAUAUUUACUU-UUAUACUU---------UCUCUUA--C-U .....((((((((.........)))((((((((.((.((....)).)).)))))..)))....-...........))-))).....---------.......--.-. ( -20.80) >DroEre_CAF1 2638 104 + 1 ACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUAUACUAUAUAUUUUU-CUAUACUUUUCGAACCUUCUCCUU--GCG ..........(((.........)))((((((((.((.((....)).)).)))))..)))..................-........................--... ( -20.70) >DroMoj_CAF1 4174 94 + 1 ACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUU-UCUAUAUUUACUUUUUAUACUU---------UCUCUUU--C-U .....((((((((.........)))((((((((.((.((....)).)).)))))..)))....-............))))).....---------.......--.-. ( -20.90) >DroAna_CAF1 3192 106 + 1 ACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUAUACUAUAUAUUUUU-CUAUACUUUUCGAACCUUCUCUUUUUCAG ..........(((.........)))((((((((.((.((....)).)).)))))..)))..................-............................. ( -20.70) >consensus ACAAUUAAAAGCGGCCCAAACACGCGCCACCGCUGUCGCUGCUGCAGCUGCGGUAUGGCUAUA_ACUAUAUAUACUU_CUAUACUU_________UCUCUUU__C_G ..........(((.........)))((((((((.((.((....)).)).)))))..)))................................................ (-20.70 = -20.70 + -0.00)

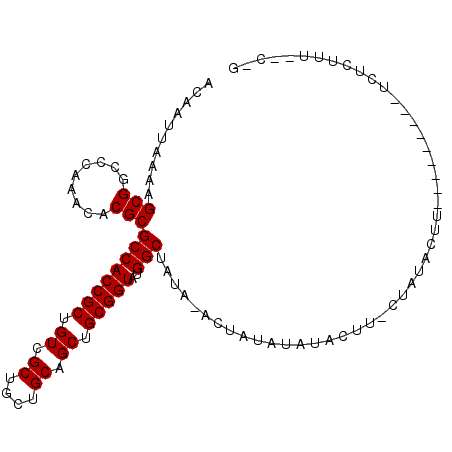
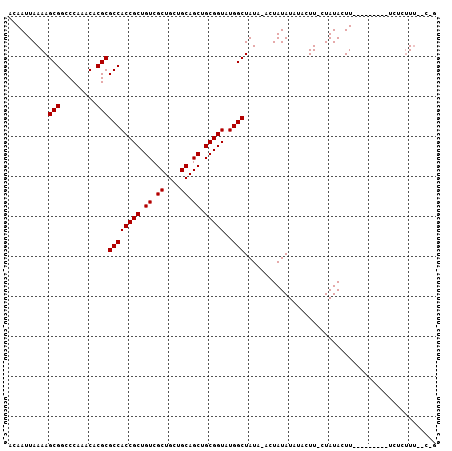
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:50 2006