
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,366,206 – 2,366,325 |
| Length | 119 |
| Max. P | 0.994006 |

| Location | 2,366,206 – 2,366,325 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.72 |
| Mean single sequence MFE | -32.62 |
| Consensus MFE | -27.30 |
| Energy contribution | -27.54 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
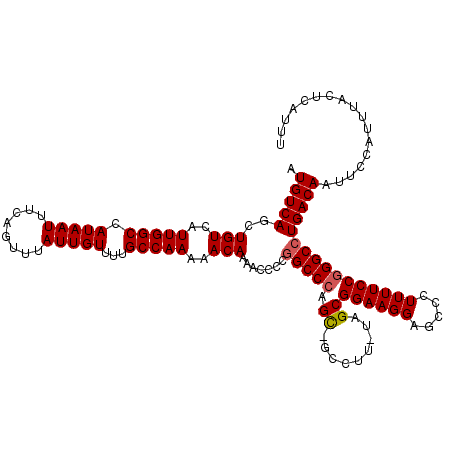
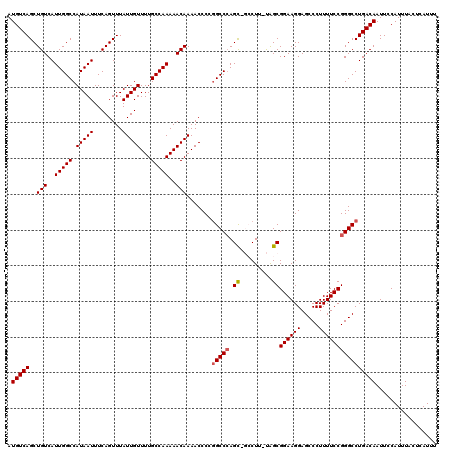
>2R_DroMel_CAF1 2366206 119 + 20766785 AUGUCAGCUGUCAUUGGCCAUAAUUUCAGUUUAUUGUUUUGCCAAAAACAAAACCCCGGCCCAGCCGCCUU-UAGCGGAAGGAGCCCUUUUCCGGGCCUGACAAUUCCAUUUACUCAUUU .(((((..(((..(((((.(((((........)))))...)))))..))).......(((((..((((...-..))))..((((.....))))))))))))))................. ( -34.30) >DroSec_CAF1 15698 119 + 1 AUGUCAGCUGUCAUUGGCCAUAAUUUCAGUUUAUUGUUUUGCCAAAAACAAAACCCCGGCCGAGCAGCCUU-UAGCGGAAGGAGCCCUUUUCCGGGCCUGACAAUUCCAUUUACUCAUUU .((((((((((..((((((.(((.......)))(((((((....)))))))......))))))))))((((-(....))))).((((......))))))))))................. ( -33.80) >DroSim_CAF1 15820 119 + 1 AUGUCAGCUGUCAUUGGCCAUAAUUUCAGUUUAUUGUUUUGCCAAAAACAAAACCCCGGCCCAGCAGCCUU-UAGCGGAAGGAGCCCUUUUCCGGGCCUGACAAUUCCAUUUACUCAUUU .(((((..(((..(((((.(((((........)))))...)))))..))).......(((((.((......-..))((((((.....))))))))))))))))................. ( -31.50) >DroEre_CAF1 15290 116 + 1 AUGUCAGCUGUCAUUGGCCAUAAUUUCAGUUUAUUGUUUUGCCAAAAACAAAACCCGGGCCCAGU---CUU-CAGCGGAAGGAGCCCUUUUCCGGGCCUGACAAUUCCAUUUACUCAUUU .(((((((((..((((....))))..)))))..(((((((....))))))).....((((((.((---...-..))((((((.....))))))))))))))))................. ( -29.60) >DroYak_CAF1 15072 117 + 1 AUGUCAGCUGUCAUUGGCCAUAAUUUCAGUUUAUUGUUUUGCCAAAAACAAAACCCAAGCCCAGC---CUUUUGGCGGAAGGAGCCCUUUUCCGGGCCUGACAAUUCCAUUUACUCAUUU .((((((.(((..(((((.(((((........)))))...)))))..)))........((((.((---(....)))((((((.....))))))))))))))))................. ( -33.90) >consensus AUGUCAGCUGUCAUUGGCCAUAAUUUCAGUUUAUUGUUUUGCCAAAAACAAAACCCCGGCCCAGC_GCCUU_UAGCGGAAGGAGCCCUUUUCCGGGCCUGACAAUUCCAUUUACUCAUUU .(((((..(((..(((((.(((((........)))))...)))))..))).......(((((.((.........))((((((.....))))))))))))))))................. (-27.30 = -27.54 + 0.24)

| Location | 2,366,206 – 2,366,325 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.72 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -30.22 |
| Energy contribution | -30.46 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
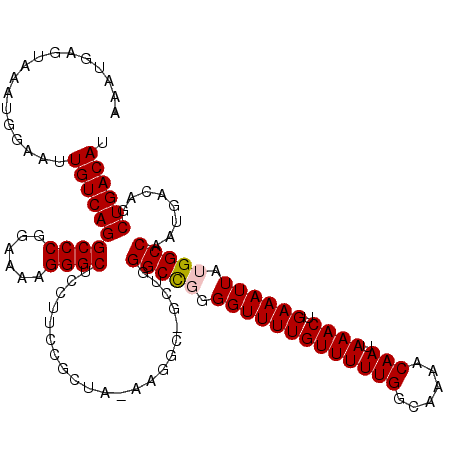
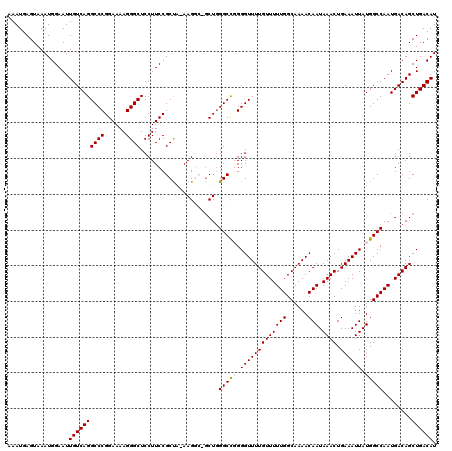
>2R_DroMel_CAF1 2366206 119 - 20766785 AAAUGAGUAAAUGGAAUUGUCAGGCCCGGAAAAGGGCUCCUUCCGCUA-AAGGCGGCUGGGCCGGGGUUUUGUUUUUGGCAAAACAAUAAACUGAAAUUAUGGCCAAUGACAGCUGACAU .................((((((((((......))))..(..((((..-...))))..)(((((..((((((((((((......))).)))).)))))..)))))........)))))). ( -38.10) >DroSec_CAF1 15698 119 - 1 AAAUGAGUAAAUGGAAUUGUCAGGCCCGGAAAAGGGCUCCUUCCGCUA-AAGGCUGCUCGGCCGGGGUUUUGUUUUUGGCAAAACAAUAAACUGAAAUUAUGGCCAAUGACAGCUGACAU .................((((((.(((((....((((.((((......-))))..))))..)))))...(((((.(((((......((((.......)))).))))).))))))))))). ( -36.60) >DroSim_CAF1 15820 119 - 1 AAAUGAGUAAAUGGAAUUGUCAGGCCCGGAAAAGGGCUCCUUCCGCUA-AAGGCUGCUGGGCCGGGGUUUUGUUUUUGGCAAAACAAUAAACUGAAAUUAUGGCCAAUGACAGCUGACAU .................((((.(((((......)))))..........-..((((((..(((((..((((((((((((......))).)))).)))))..)))))...).))))))))). ( -36.30) >DroEre_CAF1 15290 116 - 1 AAAUGAGUAAAUGGAAUUGUCAGGCCCGGAAAAGGGCUCCUUCCGCUG-AAG---ACUGGGCCCGGGUUUUGUUUUUGGCAAAACAAUAAACUGAAAUUAUGGCCAAUGACAGCUGACAU ..........(((..(((((((((((.......((((((((((....)-)))---...))))))..(((((((.....)))))))................))))..)))))).)..))) ( -32.90) >DroYak_CAF1 15072 117 - 1 AAAUGAGUAAAUGGAAUUGUCAGGCCCGGAAAAGGGCUCCUUCCGCCAAAAG---GCUGGGCUUGGGUUUUGUUUUUGGCAAAACAAUAAACUGAAAUUAUGGCCAAUGACAGCUGACAU ......((...((..((((((((((((((.....(((.......))).....---.))))))))))(((((((.....)))))))...................))))..))....)).. ( -33.40) >consensus AAAUGAGUAAAUGGAAUUGUCAGGCCCGGAAAAGGGCUCCUUCCGCUA_AAGGC_GCUGGGCCGGGGUUUUGUUUUUGGCAAAACAAUAAACUGAAAUUAUGGCCAAUGACAGCUGACAU .................((((((((((......))))......................(((((.(((((((((((((......))).)))).)))))).)))))........)))))). (-30.22 = -30.46 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:20 2006