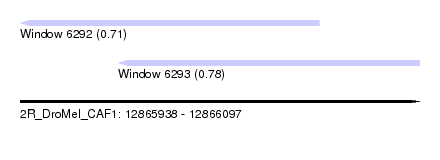
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,865,938 – 12,866,097 |
| Length | 159 |
| Max. P | 0.778227 |

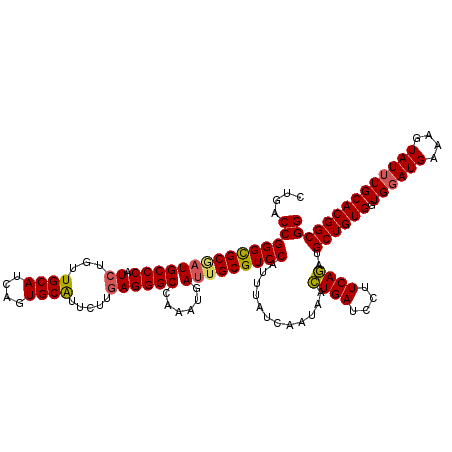
| Location | 12,865,938 – 12,866,057 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.79 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -29.23 |
| Energy contribution | -29.84 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12865938 119 - 20766785 UUGAGGGCCAAAUGAUUGCGUCCAUUUAUCAAUAACUGAUCCUUCAGAUGCUGUGGUGGAUGAAAGUAUUUGCACGGCGGUUUAUCACGAUUACAGCGGCCCUGCAAGUAAAUUCAA-GC ((((((((((((((........)))))........((((....))))..(((((((((((((((.((.........))..)))))).).)))))))))))))).)))..........-.. ( -33.80) >DroSim_CAF1 221211 111 - 1 UUGAGGGCCAAAUGA-UGCGUCCAU-UAUC-AUAACUGAUCCUUCAGAUGCUGUG-UGGAUGAAAGUAU-UGCACGGCGGU-UAUCACGA-UACAGCGGCCCUGCAAGAAAA-UCAA-GU (((((((((..((((-((.......-))))-))..((((....))))..((((((-(((((((..((..-......))..)-)))).).)-)))))))))))).))).....-....-.. ( -35.70) >DroEre_CAF1 213330 119 - 1 UUGAGGGCCAAAUGAUUGCGUCCAUUUAUCAAUAACUGAUCCUUCAGAUGCUGUGGUGGAUGAAAGUAUUUGCACGGCGGUUUAUCACGAUUACAGCGGCCCUGCAAGAAAAUUCAA-AG ((((((((((((((........)))))........((((....))))..(((((((((((((((.((.........))..)))))).).)))))))))))))).)))..........-.. ( -33.80) >DroWil_CAF1 320797 120 - 1 UUUAGGGCCAAAUGAUUGCGUCCAUUUAUCAAUAAUUGAUCCUUCAAAUGCUGUGGUGGAUGAAAAUAUUUGCACGGCGGUUUAUCACGAUUACAGCGACCCUGCAAAAGUAAGAAAGAA ..(((((.((((((........)))))........((((....))))..((((((((.(.(((.((...((((...)))).)).)))).))))))))).)))))................ ( -23.80) >DroYak_CAF1 219331 119 - 1 UUGAGGGCCAAAUGAUUGCGUCCAUUUAUCAAUAACUGAUCCUUCAGAUGCUGUGGUGGAUGAAAGUAUUUGCACGGCGGUUUAUCACGAUUACAGCGGCCCUGCAAGAAAAUUCAA-AC ((((((((((((((........)))))........((((....))))..(((((((((((((((.((.........))..)))))).).)))))))))))))).)))..........-.. ( -33.80) >consensus UUGAGGGCCAAAUGAUUGCGUCCAUUUAUCAAUAACUGAUCCUUCAGAUGCUGUGGUGGAUGAAAGUAUUUGCACGGCGGUUUAUCACGAUUACAGCGGCCCUGCAAGAAAAUUCAA_AC ((((((((((((((........)))))........((((....))))..((((((((.(.(((.((...((((...)))).)).)))).)))))))))))))).)))............. (-29.23 = -29.84 + 0.61)

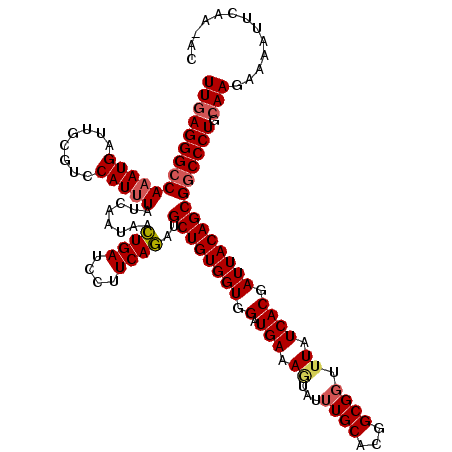
| Location | 12,865,977 – 12,866,097 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.42 |
| Mean single sequence MFE | -38.44 |
| Consensus MFE | -35.26 |
| Energy contribution | -35.30 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12865977 120 - 20766785 UUGGCCGGGCGCGAUGCCCAUCUGUUGCAUCAGUGCAUUCUUGAGGGCCAAAUGAUUGCGUCCAUUUAUCAAUAACUGAUCCUUCAGAUGCUGUGGUGGAUGAAAGUAUUUGCACGGCGG ....(((((((((((((((.((...((((....)))).....))))))......)))))))))............((((....))))..(((((.(..((((....))))..)))))))) ( -40.00) >DroSim_CAF1 221247 115 - 1 UUGGCCGGGCGCGAUGCCCAUCUGUUGCAUCAGUGCAUUCUUGAGGGCCAAAUGA-UGCGUCCAU-UAUC-AUAACUGAUCCUUCAGAUGCUGUG-UGGAUGAAAGUAU-UGCACGGCGG ((((((((((.....)))).((...((((....)))).....)).))))))((((-((.......-))))-))..((((....)))).(((((((-..(((....)).)-..))))))). ( -40.30) >DroEre_CAF1 213369 120 - 1 CUGACCGGGCGCGAUGCCCAUCUGUUGCAUCAGUGCAUUCUUGAGGGCCAAAUGAUUGCGUCCAUUUAUCAAUAACUGAUCCUUCAGAUGCUGUGGUGGAUGAAAGUAUUUGCACGGCGG ....(((((((((((((((.((...((((....)))).....))))))......)))))))))............((((....))))..(((((.(..((((....))))..)))))))) ( -40.30) >DroWil_CAF1 320837 120 - 1 CUGACCGGGUGCUAUGCCCAUCUGUUGCAUCAAUGCGUUCUUUAGGGCCAAAUGAUUGCGUCCAUUUAUCAAUAAUUGAUCCUUCAAAUGCUGUGGUGGAUGAAAAUAUUUGCACGGCGG ....(((.((((...((((......((((....)))).......)))).(((((.((.((((((((...((.((.((((....)))).)).)).)))))))).)).)))))))))..))) ( -31.02) >DroYak_CAF1 219370 120 - 1 CUGACCGGGCGCAAUGCCCAUCUGUUGCAUCAGUGCAUUCUUGAGGGCCAAAUGAUUGCGUCCAUUUAUCAAUAACUGAUCCUUCAGAUGCUGUGGUGGAUGAAAGUAUUUGCACGGCGG ....(((((((((((((((.((...((((....)))).....))))))......)))))))))............((((....))))..(((((.(..((((....))))..)))))))) ( -40.60) >consensus CUGACCGGGCGCGAUGCCCAUCUGUUGCAUCAGUGCAUUCUUGAGGGCCAAAUGAUUGCGUCCAUUUAUCAAUAACUGAUCCUUCAGAUGCUGUGGUGGAUGAAAGUAUUUGCACGGCGG ....(((((((((((((((.((...((((....)))).....))))))......)))))))))............((((....))))..((((((.((((((....)))))))))))))) (-35.26 = -35.30 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:44 2006