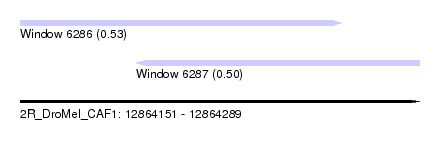
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 12,864,151 – 12,864,289 |
| Length | 138 |
| Max. P | 0.531691 |

| Location | 12,864,151 – 12,864,262 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.42 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -15.08 |
| Energy contribution | -14.64 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
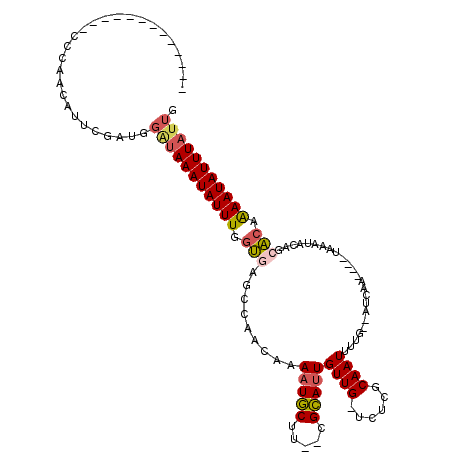
>2R_DroMel_CAF1 12864151 111 + 20766785 GUUUCGCACAGAAGGAUGGAUAAAAAUAUUUGCUUGUCGUCAAUAAAUAUUUUGUGCUGUAUUUA----UUGAU--CAAAAUUGCGAGG-CAACAAUGCA--AAGCAUUUUGUUGGCUCA .....(((......((..(.((((....)))).)..))((((((((((((........)))))))----)))))--......)))(((.-(((((((((.--..))))..))))).))). ( -28.40) >DroSim_CAF1 219438 111 + 1 GUUUCGCACAGAAGGAUGGAUAAAAAUAUUUGCUUGUCGUCAAUAAAUAUUUUGUGCUGUAUUUA----UUGAU--CAAAAUUGCGAGG-CAACAAUGCA--AAGCAUUUUGUUGGCUCA .....(((......((..(.((((....)))).)..))((((((((((((........)))))))----)))))--......)))(((.-(((((((((.--..))))..))))).))). ( -28.40) >DroEre_CAF1 211581 111 + 1 GUUUCGCACAGAAGGAUGGAUAAAAAUAUUUGCUUGUCGUCAAUAAAUAUUCUGUGCUGUAUUUA----UUGAU--CAAAAUUGCGAGA-CAACAAUGCG--AAGCAUUUCGUUGGCUAA ((((((((......((..(.((((....)))).)..))((((((((((((........)))))))----)))))--......)))))))-)..(((((.(--(....)).)))))..... ( -26.50) >DroWil_CAF1 318067 112 + 1 UUUUUGAGUUUAAGCA--GCCAAAAAUAUUUGCUUAUUGUCAAUAAAUAUUUUGC-AUGCAUUUUGCAUUUGAUAGCAAAAUUGCAAAACCAACAUUACCAAAAGUAUUU-AUU----AG .....((...((((((--(..........)))))))...))((((((((((((..-.((((((((((........)))))).))))...............)))))))))-)))----.. ( -19.87) >DroYak_CAF1 217557 111 + 1 GUUUCGCACAGAAGGAUGGAUAAAAAUAUUUGCUUGUCGUCAAUAAAUAUUUUGUGCUGUAUUUA----UUGAU--CAAAAUUGCGAGA-CAACAAUGCG--AAGCAUUUUGUUGGUUCA ((((((((......((..(.((((....)))).)..))((((((((((((........)))))))----)))))--......)))))))-)........(--((.((......)).))). ( -28.10) >consensus GUUUCGCACAGAAGGAUGGAUAAAAAUAUUUGCUUGUCGUCAAUAAAUAUUUUGUGCUGUAUUUA____UUGAU__CAAAAUUGCGAGA_CAACAAUGCA__AAGCAUUUUGUUGGCUCA .((((((((((...........((((((((((.(((....)))))))))))))...)))).........(((....)))....)))))).(((((((((.....)))))..))))..... (-15.08 = -14.64 + -0.44)


| Location | 12,864,191 – 12,864,289 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.15 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -10.12 |
| Energy contribution | -10.60 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 12864191 98 - 20766785 -------------CCCAACGUUCGAUGGAUAAAUAUUUGGUGAGCCAACAAAAUGCUU--UGCAUUGUUG-CCUCGCAAUUUUG--AUCAA----UAAAUACAGCACAAAAUAUUUAUUG -------------.(((.(....).)))((((((((((.(((((.(((((..((((..--.)))))))))-.))))).((((..--.....----.))))........)))))))))).. ( -20.20) >DroSim_CAF1 219478 98 - 1 -------------CCCAACGUUCGAUGGAUAAAUAUUUGGUGAGCCAACAAAAUGCUU--UGCAUUGUUG-CCUCGCAAUUUUG--AUCAA----UAAAUACAGCACAAAAUAUUUAUUG -------------.(((.(....).)))((((((((((.(((((.(((((..((((..--.)))))))))-.))))).((((..--.....----.))))........)))))))))).. ( -20.20) >DroEre_CAF1 211621 98 - 1 -------------CCCAUCAUUCGAUGGGUAAAUAUUUGGUUAGCCAACGAAAUGCUU--CGCAUUGUUG-UCUCGCAAUUUUG--AUCAA----UAAAUACAGCACAGAAUAUUUAUUG -------------((((((....))))))(((((((((((....))..((((((((..--.(((....))-)...)).))))))--.....----.............)))))))))... ( -21.60) >DroWil_CAF1 318105 112 - 1 AAUGUUUCUCAUUUUGAAAAUUGGCUG--AAAAUAUUUGACU----AAU-AAAUACUUUUGGUAAUGUUGGUUUUGCAAUUUUGCUAUCAAAUGCAAAAUGCAU-GCAAAAUAUUUAUUG ....((((.......))))....((..--(((.((((((...----..)-))))).)))..))((((...(((((((((((((((........)))))))...)-)))))))...)))). ( -21.90) >DroYak_CAF1 217597 97 - 1 -------------C-CAUCAUUCGAUGGAUAAAUAUUUGGUGAACCAACAAAAUGCUU--CGCAUUGUUG-UCUCGCAAUUUUG--AUCAA----UAAAUACAGCACAAAAUAUUUAUUG -------------(-((((....)))))((((((((((.((((..(((((..((((..--.)))))))))-..)))).((((..--.....----.))))........)))))))))).. ( -21.60) >consensus _____________CCCAACAUUCGAUGGAUAAAUAUUUGGUGAGCCAACAAAAUGCUU__CGCAUUGUUG_UCUCGCAAUUUUG__AUCAA____UAAAUACAGCACAAAAUAUUUAUUG ...........................(((((((((((.(((.........(((((.....)))))((((......))))........................))).))))))))))). (-10.12 = -10.60 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:06:38 2006